A transformer-based weakly supervised computational pathology method for clinical-grade diagnosis and molecular marker discovery of gliomas
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
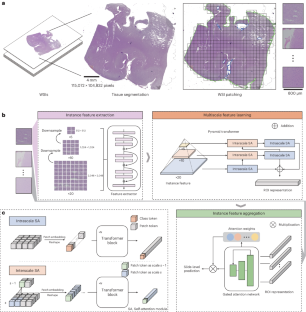
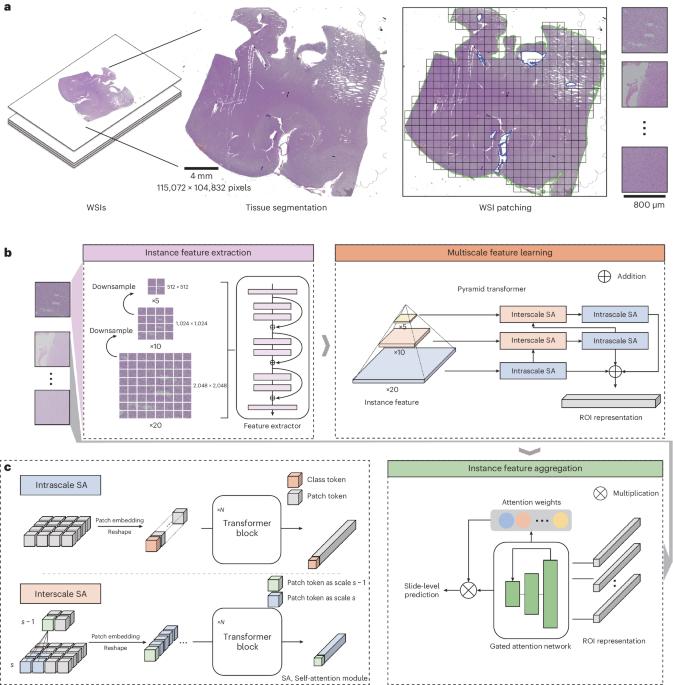
The complex diagnostic criteria for gliomas pose great challenges for making accurate diagnoses with computational pathology methods. There are no in-depth analyses of the accuracy, reliability and auxiliary capability of present approaches from a clinical perspective. Previous studies have overlooked the exploration of molecular and morphological correlations. To overcome these limitations, we propose ROAM, a multiple-instance learning model based on large regions of interest and a pyramid transformer. ROAM enlarges regions of interest to facilitate the consideration of tissue contexts. It utilizes the pyramid transformer to model both intrascale and interscale correlations of morphological features and leverages class-specific multiple-instance learning based on attention to extract slide-level visual representations that can be used to diagnose gliomas. Through comprehensive experiments on both in-house and external glioma datasets, we demonstrate that ROAM can automatically capture key morphological features consistent with the experience of pathologists and thus provide accurate, reliable and adaptable clinical-grade diagnoses of gliomas. Moreover, ROAM has clinical value for auxiliary diagnoses and could pave the way for the study of molecular and morphological correlations. ROAM, based on large regions of interest and a pyramid transformer, can automatically capture key morphological features consistent with the experience of pathologists to provide accurate, reliable and adaptable clinical-grade diagnoses of gliomas while advancing the discovery of molecular and morphological markers related to glioma diagnosis.
基于变换器的弱监督计算病理学方法,用于胶质瘤的临床分级诊断和分子标记物发现
神经胶质瘤的诊断标准十分复杂,这给利用计算病理学方法做出准确诊断带来了巨大挑战。目前还没有从临床角度对现有方法的准确性、可靠性和辅助能力进行深入分析。以往的研究忽略了对分子和形态学相关性的探讨。为了克服这些局限性,我们提出了基于大兴趣区域和金字塔转换器的多实例学习模型 ROAM。ROAM 扩大了感兴趣区域,便于考虑组织背景。它利用金字塔变换器对形态特征的级内和级间相关性进行建模,并利用基于注意力的特定类别多实例学习来提取可用于诊断胶质瘤的幻灯片级视觉表征。通过对内部和外部胶质瘤数据集的全面实验,我们证明 ROAM 可以自动捕捉与病理学家经验一致的关键形态特征,从而提供准确、可靠和适应性强的胶质瘤临床级诊断。此外,ROAM 还具有辅助诊断的临床价值,可为分子和形态学相关性研究铺平道路。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


