Revealing the novel metabolism-related genes in the ossification of the ligamentum flavum based on whole transcriptomic data
Abstract
Backgrounds
The ossification of the ligamentum flavum (OLF) is one of the major causes of thoracic myelopathy. Previous studies indicated there might be a potential link between metabolic disorder and pathogenesis of OLF. The aim of this study was to determine the potential role of metabolic disorder in the pathogenesis of OLF using the strict bioinformatic workflow for metabolism-related genes and experimental validation.
Methods
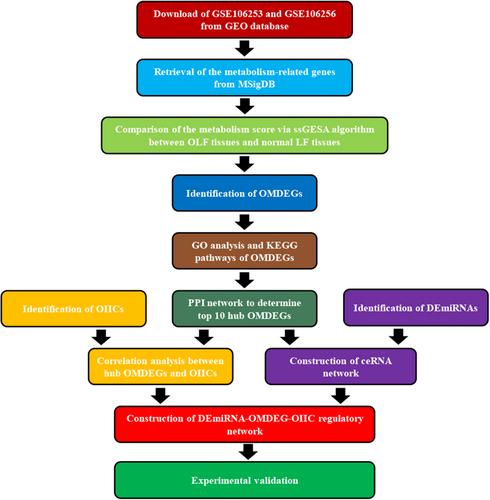
A series of bioinformatic approaches based on metabolism-related genes were conducted to compare the metabolism score between OLF tissues and normal ligamentum flavum (LF) tissues using the single sample gene set enrichment analysis. The OLF-related and metabolism-related differentially expressed genes (OMDEGs) were screened out, and the biological functions of OMDEGs were explored, including the Gene Ontology enrichment analysis, Kyoto Encyclopedia of Genes and Genomes enrichment analysis, and protein–protein interaction. The competing endogenous RNA (ceRNA) network based on pairs of miRNA-hub OMDEGs was constructed. The correlation analysis was conducted to explore the potential relationship between metabolic disorder and immunity abnormality in OLF. In the end, the cell experiments were performed to validate the roles of GBE1 and TNF-α in the osteogenic differentiation of LF cells.
Results
There was a significant difference of metabolism score between OLF tissues and normal LF tissues. Forty-nine OMDEGs were screened out and their biological functions were determined. The ceRNA network containing three hub OMDEGs and five differentially expressed miRNAs (DEmiRNAs) was built. The correlation analysis between hub OMDEGs and OLF-related infiltrating immune cells indicated that metabolic disorder might contribute to the OLF via altering the local immune status of LF tissues. The cell experiments determined the important roles of GBE1 expression and TNF-α in the osteogenic differentiation of LF cells.
Conclusions
This research, for the first time, preliminarily illustrated the vital role of metabolic disorder in the pathogenesis of OLF using strict bioinformatic algorithms and experimental validation for metabolism-related genes, which could provide new insights for investigating disease mechanism and screening effective therapeutic targets of OLF in the future.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


