An interpretable deep learning framework for genome-informed precision oncology
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
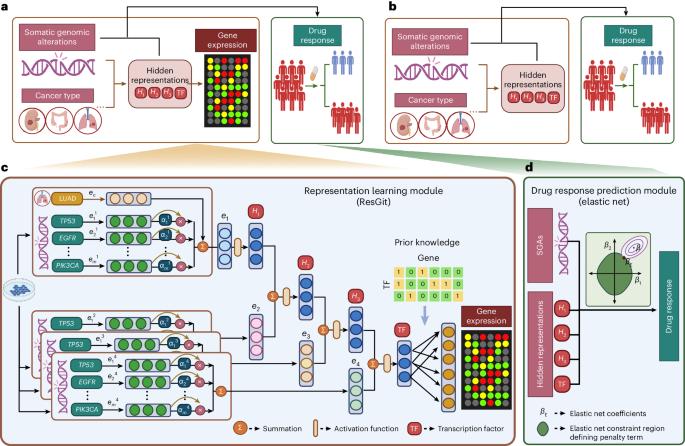
Cancers result from aberrations in cellular signalling systems, typically resulting from driver somatic genome alterations (SGAs) in individual tumours. Precision oncology requires understanding the cellular state and selecting medications that induce vulnerability in cancer cells under such conditions. To this end, we developed a computational framework consisting of two components: (1) a representation-learning component, which learns a representation of the cellular signalling systems when perturbed by SGAs and uses a biologically motivated and interpretable deep learning model, and (2) a drug-response prediction component, which predicts drug responses by leveraging the information of the cellular state of the cancer cells derived by the first component. Our cell-state-oriented framework notably improves the accuracy of predictions of drug responses compared to models using SGAs directly in cell lines. Moreover, our model performs well with real patient data. Importantly, our framework enables the prediction of responses to chemotherapy agents based on SGAs, thus expanding genome-informed precision oncology beyond molecularly targeted drugs. Precision oncology requires analysis of genomic alterations in cancer cells. Ren et al. develop an interpretable artificial intelligence framework that transforms somatic genomic alterations into representations of cellular signalling systems and accurately predicts cells’ responses to anticancer drugs.
用于基因组信息精准肿瘤学的可解释深度学习框架
癌症源于细胞信号系统的畸变,通常是由单个肿瘤中的体细胞基因组驱动改变(SGA)引起的。精准肿瘤学需要了解细胞状态,并选择能在这种条件下诱导癌细胞脆弱性的药物。为此,我们开发了一个由两部分组成的计算框架:(1)表征学习组件,学习细胞信号系统在受到 SGAs 干扰时的表征,并使用具有生物学动机和可解释的深度学习模型;(2)药物反应预测组件,利用第一个组件得出的癌细胞状态信息预测药物反应。与直接在细胞系中使用 SGA 的模型相比,我们以细胞状态为导向的框架显著提高了药物反应预测的准确性。此外,我们的模型在实际患者数据中表现良好。重要的是,我们的框架可以根据 SGA 预测化疗药物的反应,从而将基因组信息精准肿瘤学扩展到分子靶向药物之外。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


