Identification of Parkinson’s disease PACE subtypes and repurposing treatments through integrative analyses of multimodal data
IF 12.4
1区 医学
Q1 HEALTH CARE SCIENCES & SERVICES
引用次数: 0
Abstract
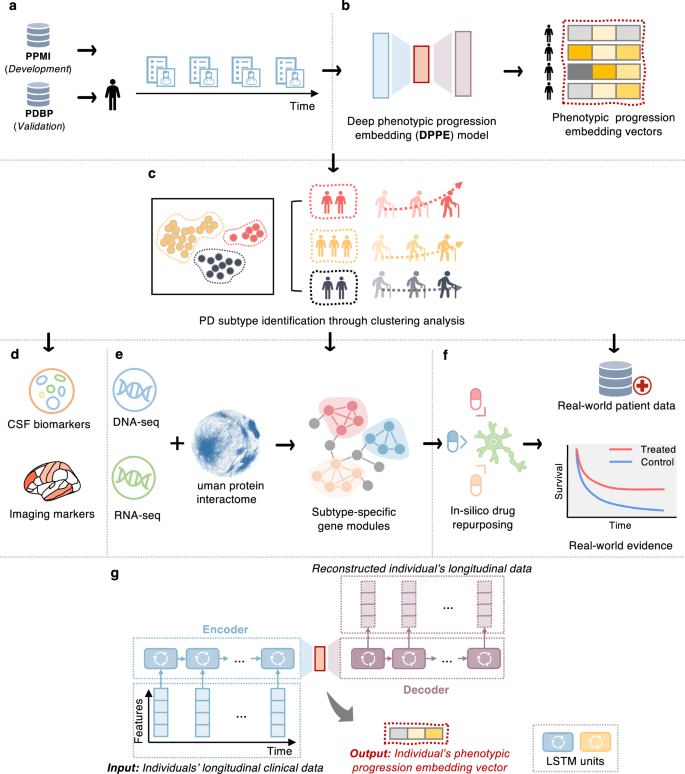
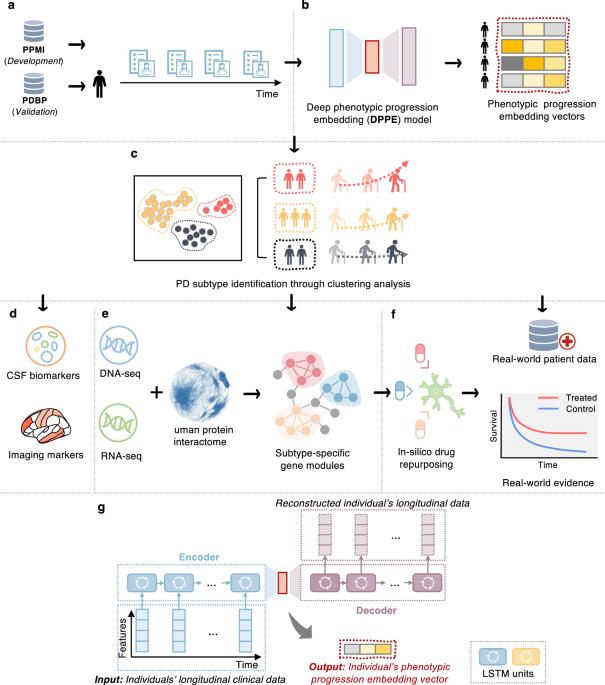
Parkinson’s disease (PD) is a serious neurodegenerative disorder marked by significant clinical and progression heterogeneity. This study aimed at addressing heterogeneity of PD through integrative analysis of various data modalities. We analyzed clinical progression data (≥5 years) of individuals with de novo PD using machine learning and deep learning, to characterize individuals’ phenotypic progression trajectories for PD subtyping. We discovered three pace subtypes of PD exhibiting distinct progression patterns: the Inching Pace subtype (PD-I) with mild baseline severity and mild progression speed; the Moderate Pace subtype (PD-M) with mild baseline severity but advancing at a moderate progression rate; and the Rapid Pace subtype (PD-R) with the most rapid symptom progression rate. We found cerebrospinal fluid P-tau/α-synuclein ratio and atrophy in certain brain regions as potential markers of these subtypes. Analyses of genetic and transcriptomic profiles with network-based approaches identified molecular modules associated with each subtype. For instance, the PD-R-specific module suggested STAT3, FYN, BECN1, APOA1, NEDD4, and GATA2 as potential driver genes of PD-R. It also suggested neuroinflammation, oxidative stress, metabolism, PI3K/AKT, and angiogenesis pathways as potential drivers for rapid PD progression (i.e., PD-R). Moreover, we identified repurposable drug candidates by targeting these subtype-specific molecular modules using network-based approach and cell line drug-gene signature data. We further estimated their treatment effects using two large-scale real-world patient databases; the real-world evidence we gained highlighted the potential of metformin in ameliorating PD progression. In conclusion, this work helps better understand clinical and pathophysiological complexity of PD progression and accelerate precision medicine.
通过对多模态数据的综合分析,确定帕金森病 PACE 亚型并重新确定治疗目标。
帕金森病(PD)是一种严重的神经退行性疾病,具有显著的临床和进展异质性。本研究旨在通过综合分析各种数据模式来解决帕金森病的异质性问题。我们利用机器学习和深度学习分析了新发帕金森病患者的临床进展数据(≥5 年),以表征个体的表型进展轨迹,从而对帕金森病进行亚型分析。我们发现了表现出不同进展模式的三种步伐型帕金森病亚型:基线严重程度较轻、进展速度较慢的步进型帕金森病亚型(PD-I);基线严重程度较轻但进展速度中等的中度步伐型帕金森病亚型(PD-M);症状进展速度最快的快速步伐型帕金森病亚型(PD-R)。我们发现脑脊液中的P-tau/α-突触核蛋白比率和某些脑区的萎缩是这些亚型的潜在标记物。利用基于网络的方法对基因和转录组图谱进行分析,发现了与每种亚型相关的分子模块。例如,PD-R 特异性模块认为 STAT3、FYN、BECN1、APOA1、NEDD4 和 GATA2 是 PD-R 的潜在驱动基因。它还认为神经炎症、氧化应激、新陈代谢、PI3K/AKT 和血管生成通路是 PD 快速进展(即 PD-R)的潜在驱动基因。此外,我们利用基于网络的方法和细胞系药物基因特征数据,通过靶向这些亚型特异性分子模块确定了可再利用的候选药物。我们利用两个大规模真实世界患者数据库进一步估算了它们的治疗效果;我们获得的真实世界证据突显了二甲双胍在改善帕金森病进展方面的潜力。总之,这项工作有助于更好地理解帕金森病进展的临床和病理生理学复杂性,加速精准医疗的发展。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

NPJ Digital Medicine
Multiple-
CiteScore
25.10
自引率
3.30%
发文量
170
审稿时长
15 weeks
期刊介绍:
npj Digital Medicine is an online open-access journal that focuses on publishing peer-reviewed research in the field of digital medicine. The journal covers various aspects of digital medicine, including the application and implementation of digital and mobile technologies in clinical settings, virtual healthcare, and the use of artificial intelligence and informatics.
The primary goal of the journal is to support innovation and the advancement of healthcare through the integration of new digital and mobile technologies. When determining if a manuscript is suitable for publication, the journal considers four important criteria: novelty, clinical relevance, scientific rigor, and digital innovation.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


