Ligandability at the Membrane Interface of GPx4 Revealed through a Reverse Micelle Fragment Screening Platform
引用次数: 0
Abstract
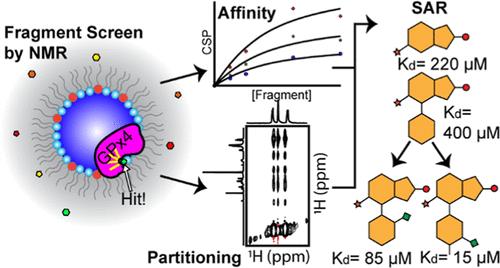
While they account for a large portion of drug targets, membrane proteins present a unique challenge for drug discovery. Peripheral membrane proteins (PMPs), a class of water-soluble proteins that bind to membranes, are also difficult targets, particularly those that function only when bound to membranes. The protein–membrane interface in PMPs is often where functional interactions and catalysis occur, making it a logical target for inhibition. However, protein–membrane interfaces are underexplored spaces in inhibitor design, and there is a need for enhanced methods for small-molecule ligand discovery. In an effort to better initiate drug discovery efforts for PMPs, this study presents a screening methodology using membrane-mimicking reverse micelles (mmRM) and NMR-based fragment screening to assess ligandability at the protein–membrane interface. The proof-of-principle target, glutathione peroxidase 4 (GPx4), is a lipid hydroperoxidase that is essential for the oxidative protection of membranes and thereby the prevention of ferroptosis. GPx4 inhibition is promising for therapy-resistant cancer therapy, but current inhibitors are generally covalent ligands with limited clinical utility. Presented here is the discovery of noncovalent small-molecule ligands for membrane-bound GPx4 revealed through the mmRM fragment screening methodology. The fragments were tested against GPx4 under bulk aqueous conditions and displayed little to no binding to the protein without embedment into the membrane. The 9 hits had varying affinities and partitioning coefficients and revealed properties of fragments that bind within the protein–membrane interface. Additionally, a secondary screen confirmed the potential to progress the fragments by enhancing the affinity from >200 to ∼15 μM with the addition of certain hydrophobic groups. This study presents an advancement of screening capabilities for membrane-associated proteins, reveals ligandability within the GPx4 protein–membrane interface, and may serve as a starting point for developing noncovalent inhibitors of GPx4.
通过反向胶束片段筛选平台揭示 GPx4 膜界面的配体性
虽然膜蛋白占药物靶点的很大一部分,但它们也给药物发现带来了独特的挑战。外周膜蛋白(PMPs)是一类与膜结合的水溶性蛋白质,也是难以发现的靶点,尤其是那些只有与膜结合时才能发挥作用的蛋白质。PMPs 中的蛋白质-膜界面通常是功能性相互作用和催化作用发生的地方,因此是一个合理的抑制靶点。然而,蛋白质-膜界面是抑制剂设计中尚未充分开发的空间,因此需要改进小分子配体的发现方法。为了更好地启动 PMPs 的药物发现工作,本研究提出了一种筛选方法,利用膜模拟反向胶束(mmRM)和基于 NMR 的片段筛选来评估蛋白质-膜界面的配体性。原理验证靶点谷胱甘肽过氧化物酶 4(GPx4)是一种脂质过氧化物酶,对膜的氧化保护至关重要,因此也是防止铁变态反应的关键。抑制 GPx4 有助于治疗耐药性癌症,但目前的抑制剂一般都是共价配体,临床应用有限。本文介绍了通过 mmRM 片段筛选方法发现的膜结合 GPx4 非共价小分子配体。这些片段在大量水溶液条件下与 GPx4 进行了测试,在没有嵌入膜的情况下,几乎没有显示出与蛋白质的结合。9 个命中的片段具有不同的亲和力和分配系数,揭示了在蛋白质-膜界面内结合的片段的特性。此外,二次筛选证实,通过添加某些疏水基团,可将片段的亲和力从 200 μM 提高到 15 μM。这项研究提高了对膜相关蛋白的筛选能力,揭示了 GPx4 蛋白-膜界面的配体性,可作为开发 GPx4 非共价抑制剂的起点。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


