Decoding dysregulated genes, molecular pathways and microRNAs involved in cervical cancer
Abstract
Background
The present study aimed to identify dysregulated genes, molecular pathways, and regulatory mechanisms in human papillomavirus (HPV)-associated cervical cancers. We have investigated the disease-associated genes along with the Gene Ontology, survival prognosis, transcription factors and the microRNA (miRNA) that are involved in cervical carcinogenesis, enabling a deeper comprehension of cervical cancer linked to HPV.
Methods
We used 10 publicly accessible Gene Expression Omnibus (GEO) datasets to examine the patterns of gene expression in cervical cancer. Differentially expressed genes (DEGs), which showed a clear distinction between cervical cancer and healthy tissue samples, were analyzed using the GEO2R tool. Additional bioinformatic techniques were used to carry out pathway analysis and functional enrichment, as well as to analyze the connection between altered gene expression and HPV infection.
Results
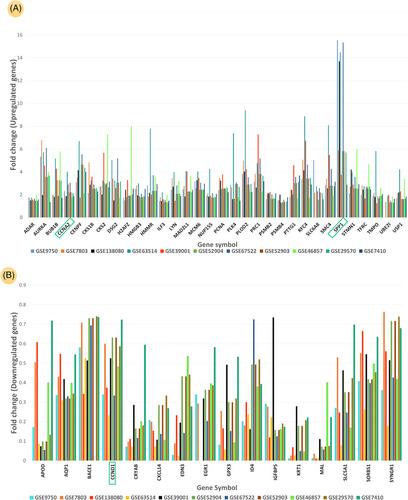
In total, 48 DEGs were identified to be differentially expressed in cervical cancer tissues in comparison to healthy tissues. Among DEGs, CCND1, CCNA2 and SPP1 were the key dysregulated genes involved in HPV-associated cervical cancer. The five common miRNAs that were identified against these genes are miR-7-5p, miR-16-5p, miR-124-3p, miR-10b-5p and miR-27a-3p. The hub-DEGs targeted by miRNA hsa-miR-27a-3p are controlled by the common transcription factor SP1.
Conclusions
The present study has identified DEGs involved in HPV-associated cervical cancer progression and the various molecular pathways and transcription factors regulating them. These findings have led to a better understanding of cervical cancer resulting in the development and identification of possible therapeutic and intervention targets, respectively.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


