Direct conformational sampling from peptide energy landscapes through hypernetwork-conditioned diffusion
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
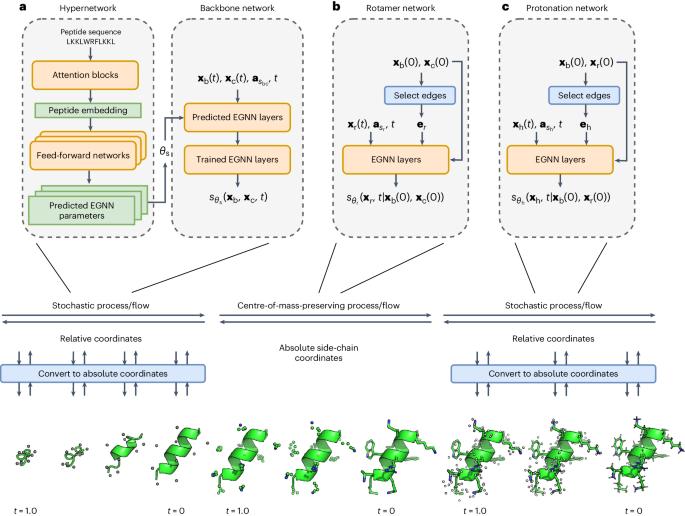
Deep learning approaches have spurred substantial advances in the single-state prediction of biomolecular structures. The function of biomolecules is, however, dependent on the range of conformations they can assume. This is especially true for peptides, a highly flexible class of molecules that are involved in numerous biological processes and are of high interest as therapeutics. Here we introduce PepFlow, a transferable generative model that enables direct all-atom sampling from the allowable conformational space of input peptides. We train the model in a diffusion framework and subsequently use an equivalent flow to perform conformational sampling. To overcome the prohibitive cost of generalized all-atom modelling, we modularize the generation process and integrate a hypernetwork to predict sequence-specific network parameters. PepFlow accurately predicts peptide structures and effectively recapitulates experimental peptide ensembles at a fraction of the running time of traditional approaches. PepFlow can also be used to sample conformations that satisfy constraints such as macrocyclization. Modelling the different structures a peptide can assume is integral to understanding their function. The authors introduce PepFlow, a sequence-conditioned deep learning model that is shown to accurately and efficiently generate peptide conformations.
通过超网络条件扩散从多肽能谱中直接进行构象采样
深度学习方法在生物分子结构的单态预测方面取得了长足进步。然而,生物分子的功能取决于它们所能呈现的构象范围。肽是一类高度灵活的分子,参与多种生物过程,作为治疗药物备受关注。在这里,我们介绍一种可转移的生成模型 PepFlow,它可以从输入肽的可构象空间中直接进行全原子采样。我们在扩散框架中训练模型,随后使用等效流进行构象采样。为了克服通用全原子建模成本过高的问题,我们将生成过程模块化,并整合了超网络来预测特定序列的网络参数。PepFlow 能准确预测多肽结构,并有效再现实验中的多肽组合,而运行时间仅为传统方法的一小部分。PepFlow 还可用于采样满足大环化等约束条件的构象。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


