Interpreting cis-regulatory mechanisms from genomic deep neural networks using surrogate models
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Deep neural networks (DNNs) have greatly advanced the ability to predict genome function from sequence. However, elucidating underlying biological mechanisms from genomic DNNs remains challenging. Existing interpretability methods, such as attribution maps, have their origins in non-biological machine learning applications and therefore have the potential to be improved by incorporating domain-specific interpretation strategies. Here we introduce SQUID (Surrogate Quantitative Interpretability for Deepnets), a genomic DNN interpretability framework based on domain-specific surrogate modelling. SQUID approximates genomic DNNs in user-specified regions of sequence space using surrogate models—simpler quantitative models that have inherently interpretable mathematical forms. SQUID leverages domain knowledge to model cis-regulatory mechanisms in genomic DNNs, in particular by removing the confounding effects that nonlinearities and heteroscedastic noise in functional genomics data can have on model interpretation. Benchmarking analysis on multiple genomic DNNs shows that SQUID, when compared to established interpretability methods, identifies motifs that are more consistent across genomic loci and yields improved single-nucleotide variant-effect predictions. SQUID also supports surrogate models that quantify epistatic interactions within and between cis-regulatory elements, as well as global explanations of cis-regulatory mechanisms across sequence contexts. SQUID thus advances the ability to mechanistically interpret genomic DNNs. The intersection of genomics and deep learning shows promise for real impact on healthcare and biological research, but the lack of interpretability in terms of biological mechanisms is limiting utility and further development. As a potential solution, Koo et al. present SQUID, an interpretability framework built using domain-specific genomic surrogate models.
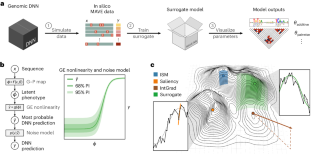
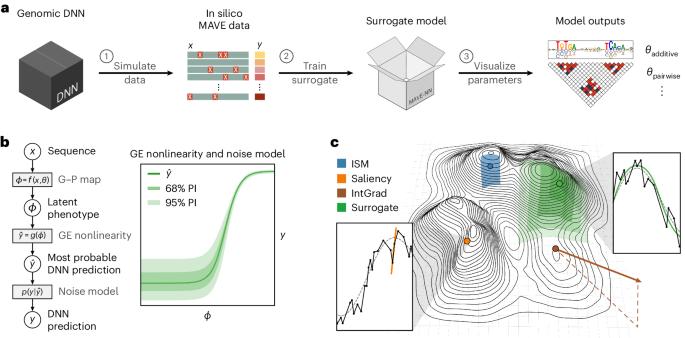
利用代用模型从基因组深度神经网络中解读顺式调控机制
深度神经网络(DNN)大大提高了从序列预测基因组功能的能力。然而,从基因组 DNNs 中阐明潜在的生物机制仍然具有挑战性。现有的可解释性方法,如归因图,起源于非生物机器学习应用,因此有可能通过纳入特定领域的解释策略来加以改进。在此,我们介绍基于特定领域代用建模的基因组 DNN 可解释性框架 SQUID(Surrogate Quantitative Interpretability for Deepnets)。SQUID 在用户指定的序列空间区域使用代用模型--具有固有可解释数学形式的更简单定量模型--来近似基因组 DNN。SQUID 利用领域知识对基因组 DNN 中的顺式调控机制进行建模,特别是消除了功能基因组学数据中的非线性和异方差噪声对模型解释的干扰效应。对多个基因组 DNN 的基准分析表明,与已有的可解释性方法相比,SQUID 能识别跨基因组位点更一致的主题,并能改进单核苷酸变异效应预测。SQUID 还支持对顺式调控元件内部和之间的表观相互作用进行量化的替代模型,以及跨序列上下文的顺式调控机制的全局解释。因此,SQUID 提高了从机理上解释基因组 DNN 的能力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


