Revealing the potential role of hub metabolism-related genes and their correlation with immune cells in acute ischemic stroke
Abstract
Objectives
Acute ischemic stroke (AIS) is caused by cerebral ischemia due to thrombosis in the blood vessel. The purpose of this study is to identify key genes related to metabolism to aid in the mechanism research and management of AIS.
Materials and Methods
Gene expression data were downloaded from the Gene Expression Omnibus database. Weighted gene co-expression network analysis, Gene Ontology and kyoto encyclopedia of genes and genomes analysis were used to identify metabolism-related genes that may be involved in the regulation of AIS. A protein protein interaction network was mapped using Cytoscape based on the STRING database. Subsequently, hub metabolism-related genes were identified based on Cytoscape-CytoNCA and Cytoscape-MCODE plug-ins. Least absolute shrinkage and selection operator algorithm and differential expression analysis. In addition, drug prediction, molecular docking, ceRNA network construction, and correlation analysis with immune cell infiltration were performed to explore their potential molecular mechanisms of action in AIS. Finally, the expression of hub gene was verified by real-time PCR.
Results
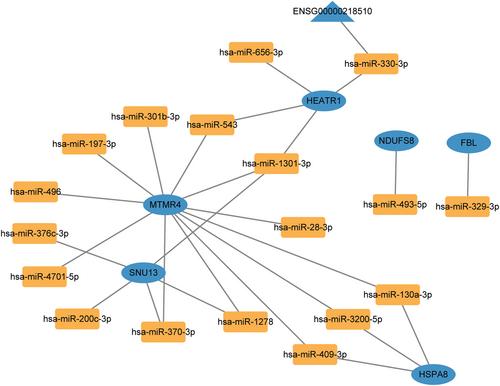
Metabolism-related genes FBL, HEATR1, HSPA8, MTMR4, NDUFC1, NDUFS8 and SNU13 were identified. The AUC values of FBL, HEATR1, HSPA8, MTMR4, NDUFS8 and SNU13 were all greater than 0.8, suggesting that they had good diagnostic accuracy. Correlation analysis found that their expression levels were also related to the infiltration levels of multiple immune cells, such as Activated.CD8.T.cell and Activated.dendritic.cell. It was found that only HSPA8 was successfully matched to drugs with literature support, and these drugs were acetaminophen, bupivacaine, dexamethasone, gentamicin, tretinoin and cisplatin. Moreover, it was also identified that the ENSG000000218510-hsa-miR-330-3p-HEATR1 axis may be involved in regulating AIS.
Conclusions
The identification of FBL, HEATR1, HSPA8, MTMR4, NDUFC1, NDUFS8 and SNU13 provides a new research direction for exploring the molecular mechanisms of AIS, which can help in clinical management and diagnosis.


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


