The structure of the TH/INS locus and the parental allele expressed are not conserved between mammals
IF 3.9
2区 生物学
Q2 ECOLOGY
引用次数: 0
Abstract
Parent-of-origin-specific expression of imprinted genes is critical for successful mammalian growth and development. Insulin, coded by the INS gene, is an important growth factor expressed from the paternal allele in the yolk sac placenta of therian mammals. The tyrosine hydroxylase gene TH encodes an enzyme involved in dopamine synthesis. TH and INS are closely associated in most vertebrates, but the mouse orthologues, Th and Ins2, are separated by repeated DNA. In mice, Th is expressed from the maternal allele, but the parental origin of expression is not known for any other mammal so it is unclear whether the maternal expression observed in the mouse represents an evolutionary divergence or an ancestral condition. We compared the length of the DNA segment between TH and INS across species and show that separation of these genes occurred in the rodent lineage with an accumulation of repeated DNA. We found that the region containing TH and INS in the tammar wallaby produces at least five distinct RNA transcripts: TH, TH-INS1, TH-INS2, lncINS and INS. Using allele-specific expression analysis, we show that the TH/INS locus is expressed from the paternal allele in pre- and postnatal tammar wallaby tissues. Determining the imprinting pattern of TH/INS in other mammals might clarify if paternal expression is the ancestral condition which has been flipped to maternal expression in rodents by the accumulation of repeat sequences.
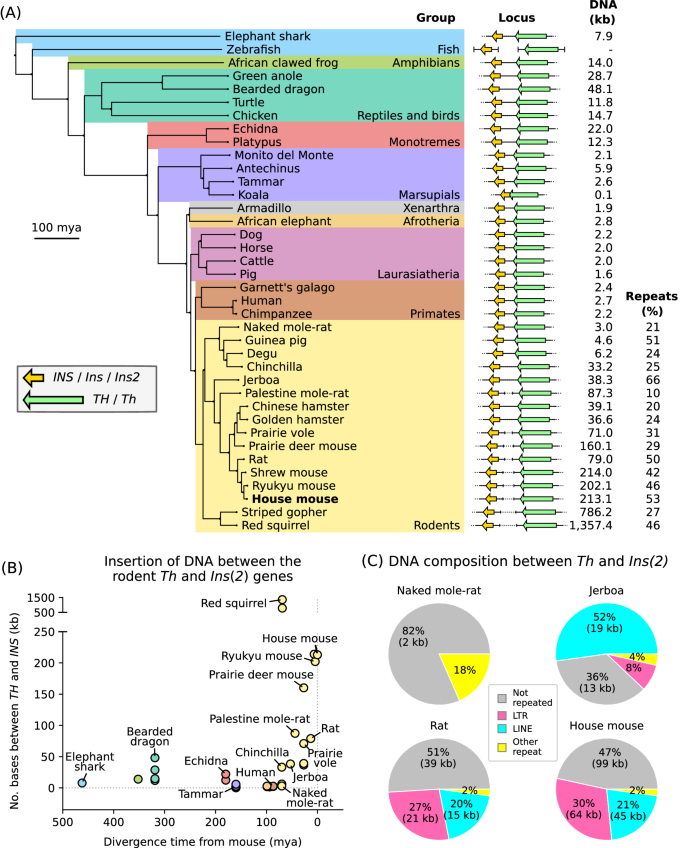
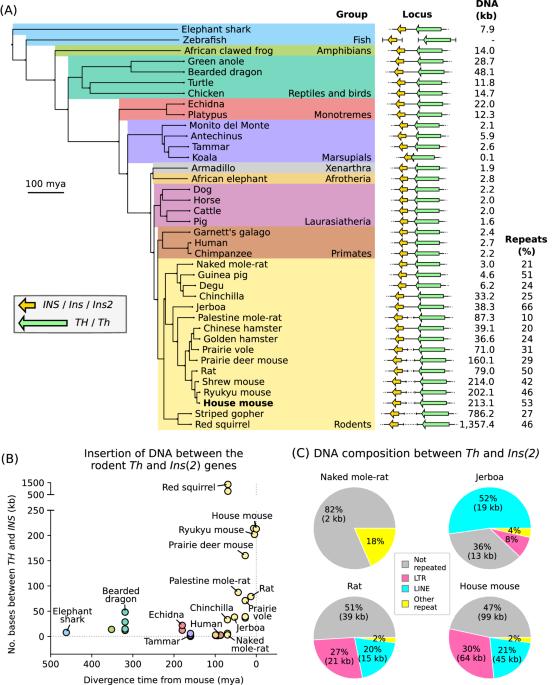
TH/INS基因座的结构和表达的亲本等位基因在哺乳动物之间并不一致。
印记基因的亲本特异性表达对哺乳动物的成功生长和发育至关重要。INS 基因编码的胰岛素是一种重要的生长因子,在有肝哺乳动物的卵黄囊胎盘中由父系等位基因表达。酪氨酸羟化酶基因 TH 编码一种参与多巴胺合成的酶。在大多数脊椎动物中,TH 和 INS 密切相关,但小鼠的同源基因 Th 和 Ins2 被重复的 DNA 分隔开来。在小鼠中,Th 是通过母系等位基因表达的,但其他哺乳动物的亲本表达起源尚不清楚,因此尚不清楚在小鼠中观察到的母系表达是代表进化分化还是祖先条件。我们比较了不同物种中 TH 和 INS 之间 DNA 区段的长度,结果表明这些基因的分离发生在啮齿动物的血统中,重复 DNA 不断积累。我们发现,在犭胥袋猴中,包含 TH 和 INS 的区域至少产生了五种不同的 RNA 转录本:TH、TH-INS1、TH-INS2、lncINS 和 INS。通过等位基因特异性表达分析,我们发现TH/INS基因座在达玛小袋鼠产前和产后组织中的表达来自父系等位基因。确定 TH/INS 在其他哺乳动物中的印记模式可能会澄清父系表达是否是啮齿类动物通过重复序列的积累而转变为母系表达的祖先条件。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


