Accurate and robust protein sequence design with CarbonDesign
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
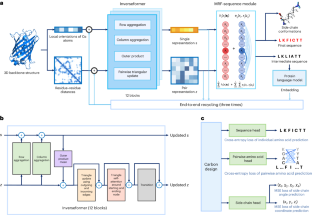
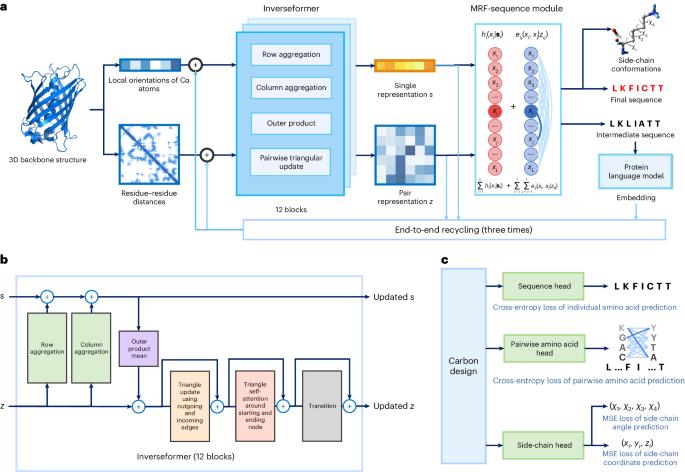
Protein sequence design is critically important for protein engineering. Despite recent advancements in deep learning-based methods, achieving accurate and robust sequence design remains a challenge. Here we present CarbonDesign, an approach that draws inspiration from successful ingredients of AlphaFold and which has been developed specifically for protein sequence design. At its core, CarbonDesign introduces Inverseformer, which learns representations from backbone structures and an amortized Markov random fields model for sequence decoding. Moreover, we incorporate other essential AlphaFold concepts into CarbonDesign: an end-to-end network recycling technique to leverage evolutionary constraints from protein language models and a multitask learning technique for generating side-chain structures alongside designed sequences. CarbonDesign outperforms other methods on independent test sets including the 15th Critical Assessment of protein Structure Prediction (CASP15) dataset, the Continuous Automated Model Evaluation (CAMEO) dataset and de novo proteins from RFDiffusion. Furthermore, it supports zero-shot prediction of the functional effects of sequence variants, making it a promising tool for applications in bioengineering. Deep learning has led to great advances in predicting protein structure from sequences. Ren and colleagues present here a method for the inverse problem of finding a sequence that results in a desired protein structure, which is inspired by various components of AlphaFold combined with Markov random fields to decode sequences more efficiently.
利用 CarbonDesign 进行精确、稳健的蛋白质序列设计
蛋白质序列设计对蛋白质工程至关重要。尽管基于深度学习的方法取得了最新进展,但实现准确、稳健的序列设计仍然是一项挑战。在此,我们介绍 CarbonDesign,这是一种从 AlphaFold 的成功要素中汲取灵感,专门为蛋白质序列设计而开发的方法。CarbonDesign 的核心是引入反演器(Inverseformer),它能从骨架结构和摊销马尔可夫随机场模型中学习序列解码的表征。此外,我们还在 CarbonDesign 中融入了 AlphaFold 的其他基本概念:利用蛋白质语言模型的进化约束的端到端网络再循环技术,以及在设计序列的同时生成侧链结构的多任务学习技术。CarbonDesign在独立测试集上的表现优于其他方法,这些测试集包括第15次蛋白质结构预测关键评估(CASP15)数据集、连续自动模型评估(CAMEO)数据集和RFDiffusion的全新蛋白质。此外,它还支持对序列变异的功能效应进行零点预测,使其成为生物工程领域的一个前景广阔的应用工具。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


