Multi-purpose RNA language modelling with motif-aware pretraining and type-guided fine-tuning
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
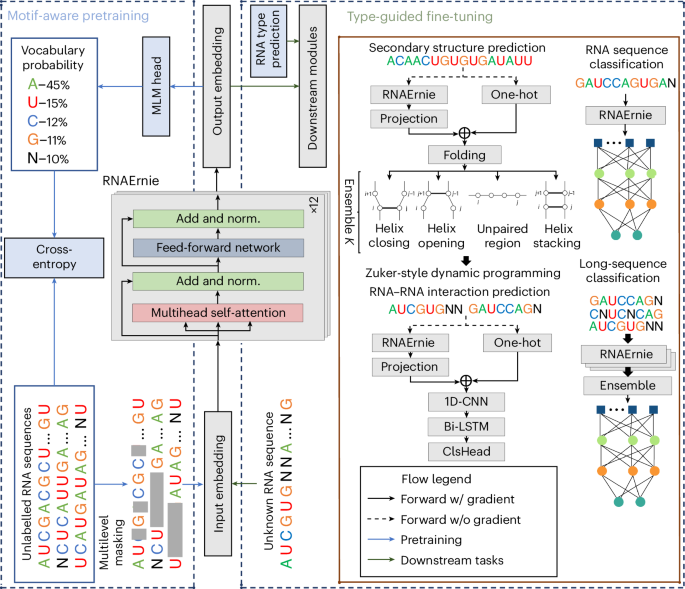
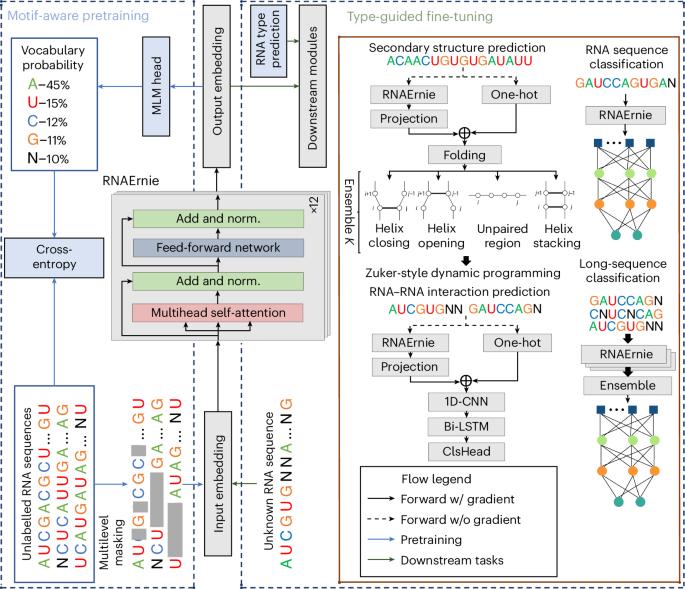
Pretrained language models have shown promise in analysing nucleotide sequences, yet a versatile model excelling across diverse tasks with a single pretrained weight set remains elusive. Here we introduce RNAErnie, an RNA-focused pretrained model built upon the transformer architecture, employing two simple yet effective strategies. First, RNAErnie enhances pretraining by incorporating RNA motifs as biological priors and introducing motif-level random masking in addition to masked language modelling at base/subsequence levels. It also tokenizes RNA types (for example, miRNA, lnRNA) as stop words, appending them to sequences during pretraining. Second, subject to out-of-distribution tasks with RNA sequences not seen during the pretraining phase, RNAErnie proposes a type-guided fine-tuning strategy that first predicts possible RNA types using an RNA sequence and then appends the predicted type to the tail of sequence to refine feature embedding in a post hoc way. Our extensive evaluation across seven datasets and five tasks demonstrates the superiority of RNAErnie in both supervised and unsupervised learning. It surpasses baselines with up to 1.8% higher accuracy in classification, 2.2% greater accuracy in interaction prediction and 3.3% improved F1 score in structure prediction, showcasing its robustness and adaptability with a unified pretrained foundation. Despite the existence of various pretrained language models for nucleotide sequence analysis, achieving good performance on a broad range of downstream tasks using a single model is challenging. Wang and colleagues develop a pretrained language model specifically optimized for RNA sequence analysis and show that it can outperform state-of-the-art methods in a diverse set of downstream tasks.
利用主题感知预训练和类型指导微调建立多用途 RNA 语言模型
预训练语言模型在分析核苷酸序列方面已显示出良好的前景,但在不同任务中使用单一预训练权重集的多功能模型仍未出现。在这里,我们介绍 RNAErnie,它是一种以 RNA 为重点的预训练模型,建立在转换器架构之上,采用了两种简单而有效的策略。首先,RNAErnie 将 RNA 主题作为生物学先验,并在基序/子序列级别的屏蔽语言建模之外引入了主题级别的随机屏蔽,从而增强了预训练效果。它还将 RNA 类型(如 miRNA、lnRNA)标记为停止词,并在预训练期间将其添加到序列中。其次,针对在预训练阶段未见过的 RNA 序列的分布外任务,RNAErnie 提出了一种类型引导的微调策略,首先使用 RNA 序列预测可能的 RNA 类型,然后将预测的类型附加到序列尾部,以事后方式完善特征嵌入。我们在七个数据集和五项任务中进行了广泛的评估,结果表明 RNAErnie 在监督和非监督学习方面都具有优势。它超越了基线,分类准确率提高了 1.8%,相互作用预测准确率提高了 2.2%,结构预测的 F1 分数提高了 3.3%,展示了它在统一预训练基础上的鲁棒性和适应性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


