Cleavage and polyadenylation machinery as a novel targetable vulnerability for human cancer
IF 4.8
3区 医学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
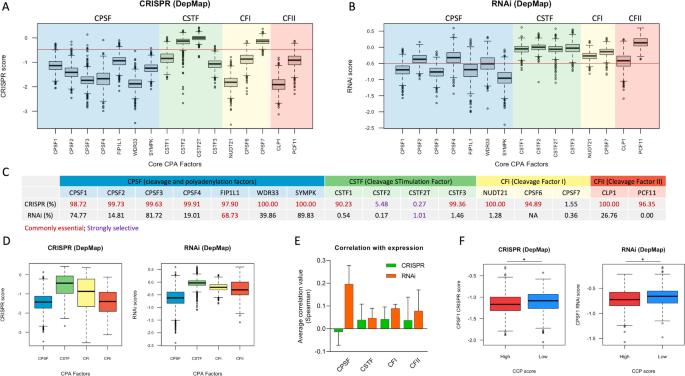
The role of alternative polyadenylation of mRNA in sustaining aggressive features of tumors is quite well established, as it is responsible for the 3’UTR shortening of oncogenes and subsequent relief from miRNA-mediated repression observed in cancer cells. However, the information regarding the vulnerability of cancer cells to the inhibition of cleavage and polyadenylation (CPA) machinery is very scattered. Only few recent reports show the antitumor activity of pharmacological inhibitors of CPSF3, one among CPA factors. More in general, the fact that deregulated CPA can be seen as a new hallmark of cancer and as a potential reservoir of novel therapeutic targets has never been formalized. Here, to extend our view on the potential of CPA inhibition (CPAi) approaches as anticancer therapies, we systematically tested the fitness of about one thousand cell lines of different cancer types upon depletion of all known CPA factors by interrogating genome-scale CRISPR and RNAi dependency maps of the DepMap project. Our analysis confirmed core and accessory CPA factors as novel vulnerabilities for human cancer, thus highlighting the potential of CPAi as anticancer therapy. Among all, CPSF1 appeared as a promising actionable candidate for drug development, as it showed low dependency scores pancancer and particularly in highly proliferating cells. In a personalized medicine perspective, the observed differential vulnerability of cancer cell lines to selected CPA factors may be used to build up signatures to predict response of individual human tumors to CPAi approaches.
裂解和多聚腺苷酸化机制是人类癌症的新型靶向漏洞
mRNA 的替代性多腺苷酸化在维持肿瘤侵袭性特征方面的作用已得到公认,因为它是导致癌基因 3'UTR 缩短以及随后摆脱癌细胞中观察到的 miRNA 介导的抑制的原因。然而,关于癌细胞易受裂解和多腺苷酸化(CPA)机制抑制的信息却非常零散。最近只有少数报道显示了 CPSF3(CPA 因子之一)药理抑制剂的抗肿瘤活性。总的来说,CPA失调可被视为癌症的新标志,也可被视为新型治疗靶点的潜在宝库,但这一事实从未得到正式确认。在这里,为了扩展我们对 CPA 抑制(CPAi)方法作为抗癌疗法的潜力的看法,我们通过询问 DepMap 项目的基因组规模 CRISPR 和 RNAi 依赖性图谱,系统地测试了约一千个不同癌症类型细胞系在去除了所有已知 CPA 因子后的适应性。我们的分析证实,核心和附属 CPA 因子是人类癌症的新漏洞,从而凸显了 CPAi 作为抗癌疗法的潜力。其中,CPSF1显示出较低的胰腺癌依赖性,尤其是在高增殖细胞中,因此有望成为可用于药物开发的候选因子。从个性化医疗的角度来看,所观察到的癌细胞系对所选 CPA 因子的不同易感性可用于建立特征,以预测个体人类肿瘤对 CPAi 方法的反应。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Cancer gene therapy
医学-生物工程与应用微生物
CiteScore
10.20
自引率
0.00%
发文量
150
审稿时长
4-8 weeks
期刊介绍:
Cancer Gene Therapy is the essential gene and cellular therapy resource for cancer researchers and clinicians, keeping readers up to date with the latest developments in gene and cellular therapies for cancer. The journal publishes original laboratory and clinical research papers, case reports and review articles. Publication topics include RNAi approaches, drug resistance, hematopoietic progenitor cell gene transfer, cancer stem cells, cellular therapies, homologous recombination, ribozyme technology, antisense technology, tumor immunotherapy and tumor suppressors, translational research, cancer therapy, gene delivery systems (viral and non-viral), anti-gene therapy (antisense, siRNA & ribozymes), apoptosis; mechanisms and therapies, vaccine development, immunology and immunotherapy, DNA synthesis and repair.
Cancer Gene Therapy publishes the results of laboratory investigations, preclinical studies, and clinical trials in the field of gene transfer/gene therapy and cellular therapies as applied to cancer research. Types of articles published include original research articles; case reports; brief communications; review articles in the main fields of drug resistance/sensitivity, gene therapy, cellular therapy, tumor suppressor and anti-oncogene therapy, cytokine/tumor immunotherapy, etc.; industry perspectives; and letters to the editor.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


