Intervertebral disc microbiome in Modic changes: Lack of result replication underscores the need for a consensus in low-biomass microbiome analysis
Abstract
Introduction
The emerging field of the disc microbiome challenges traditional views of disc sterility, which opens new avenues for novel clinical insights. However, the lack of methodological consensus in disc microbiome studies introduces discrepancies. The aims of this study were to (1) compare the disc microbiome of non-Modic (nonMC), Modic type 1 change (MC1), and MC2 discs to findings from prior disc microbiome studies, and (2) investigate if discrepancies to prior studies can be explained with bioinformatic variations.
Methods
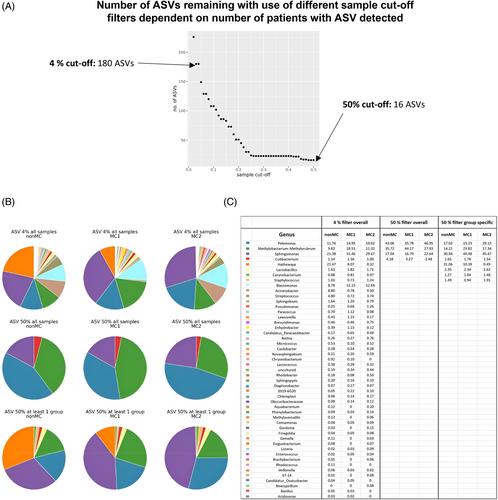
Sequencing of 16S rRNA in 70 discs (24 nonMC, 25 MC1, and 21 MC2) for microbiome profiling. The experimental setup included buffer contamination controls and was performed under aseptic conditions. Methodology and results were contrasted with previous disc microbiome studies. Critical bioinformatic steps that were different in our best-practice approach and previous disc microbiome studies (taxonomic lineage assignment, prevalence cut-off) were varied and their effect on results were compared.
Results
There was limited overlap of results with a previous study on MC disc microbiome. No bacterial genera were shared using the same bioinformatic parameters. Taxonomic lineage assignment using “amplicon sequencing variants” was more sensitive and detected 48 genera compared to 22 with “operational taxonomic units” (previous study). Increasing filter cut-off from 4% to 50% (previous study) reduced genera from 48 to 4 genera. Despite these differences, both studies observed dysbiosis with an increased abundance of gram-negative bacteria in MC discs as well as a lower beta-diversity. Cutibacterium was persistently detected in all groups independent of the bioinformatic approach, emphasizing its prevalence.
Conclusion
There is dysbiosis in MC discs. Bioinformatic parameters impact results yet cannot explain the different findings from this and a previous study. Therefore, discrepancies are likely caused by different sample preparations or true biologic differences. Harmonized protocols are required to advance understanding of the disc microbiome and its clinical implications.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


