Fluorescence resonance energy transfer at the single-molecule level
IF 56
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
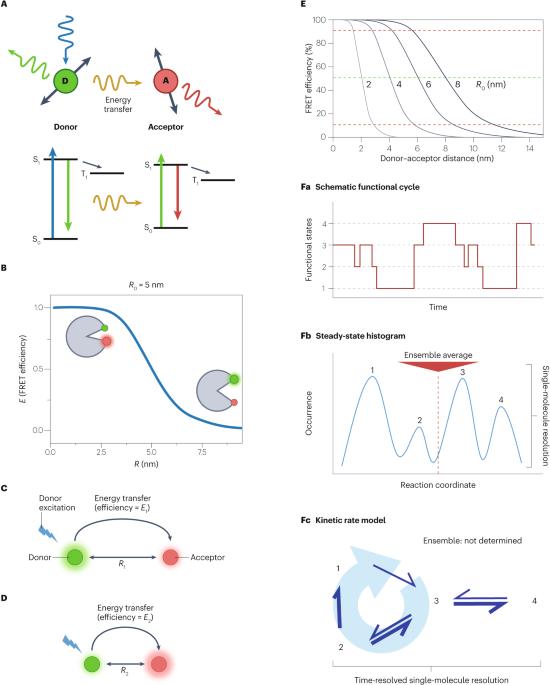
Fluorescence resonance energy transfer (FRET) is a powerful spectroscopic method for measuring distances in the 2–8 nm range. Often, conformational changes and molecular interactions are difficult or impossible to synchronize, or too rare or transient to detect using ensemble FRET. Single-molecule FRET (smFRET) opens new opportunities to probe biomolecular conformational changes or interactions that are missing in static snapshots provided by traditional structural biology tools, as well as to measure the kinetics of these dynamics on various timescales and under physiological conditions, including inside cells. Advances in labelling technologies, combining smFRET with optical and magnetic tweezers and Bayesian inference-based and information theory-based analysis tools are revealing rich biomolecular dynamics. We also discuss the challenges and opportunities in integrating dynamics into traditionally static structural biology approaches, extending smFRET into cells and tissues, advancing technical innovations and democratizing the practice of smFRET. Analysing single-molecule fluorescence resonance energy transfer (smFRET) enables an unprecedented view of the dynamics and kinetics of biomolecular conformational changes and interactions. In this Primer, Ha et al. discuss technological advances that have led to smFRET and how the method can be adapted to address various research questions in structural and molecular biology.
单分子水平的荧光共振能量转移
荧光共振能量转移(FRET)是一种功能强大的光谱方法,可用于测量 2 至 8 纳米范围内的距离。通常,构象变化和分子相互作用很难或不可能同步,或者过于罕见或瞬时,无法使用集合 FRET 进行检测。单分子 FRET(smFRET)为探测传统结构生物学工具所提供的静态快照中缺失的生物分子构象变化或相互作用提供了新的机会,也为测量这些动态在不同时间尺度和生理条件下(包括细胞内部)的动力学提供了新的机会。标记技术的进步、smFRET 与光学和磁性镊子的结合以及基于贝叶斯推理和信息论的分析工具正在揭示丰富的生物分子动力学。我们还讨论了将动力学融入传统的静态结构生物学方法、将 smFRET 扩展到细胞和组织、推进技术创新和实现 smFRET 实践民主化所面临的挑战和机遇。通过分析单分子荧光共振能量转移(smFRET),可以对生物分子构象变化和相互作用的动态和动力学有前所未有的了解。在这篇入门文章中,Ha 等人讨论了促成 smFRET 的技术进步,以及如何利用这种方法解决结构和分子生物学中的各种研究问题。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


