PocketFlow is a data-and-knowledge-driven structure-based molecular generative model
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Deep learning-based molecular generation has extensive applications in many fields, particularly drug discovery. However, the majority of current deep generative models are ligand-based and do not consider chemical knowledge in the molecular generation process, often resulting in a relatively low success rate. We herein propose a structure-based molecular generative framework with chemical knowledge explicitly considered (named PocketFlow), which generates novel ligand molecules inside protein binding pockets. In various computational evaluations, PocketFlow showed state-of-the-art performance, with generated molecules being 100% chemically valid and highly drug-like. Ablation experiments prove the critical role of chemical knowledge in ensuring the validity and drug-likeness of the generated molecules. We applied PocketFlow to two new target proteins that are related to epigenetic regulation, HAT1 and YTHDC1, and successfully obtained wet-lab validated bioactive compounds. The binding modes of the active compounds with target proteins are close to those predicted by molecular docking and further confirmed by the X-ray crystal structure. All the results suggest that PocketFlow is a useful deep generative model, capable of generating innovative bioactive molecules from scratch given a protein binding pocket. Deep learning generative approaches have been used in recent years to discover new molecules with drug-like properties. To improve the performance of such approaches, Yang et al. add chemical binding knowledge to a deep generative framework and demonstrate, including by wet-lab verification, that the method can find valid molecules that successfully bind to target proteins.
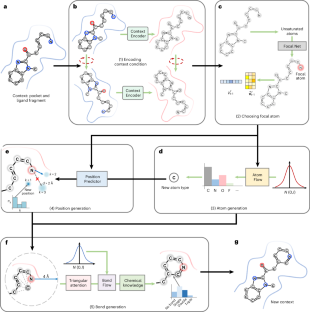
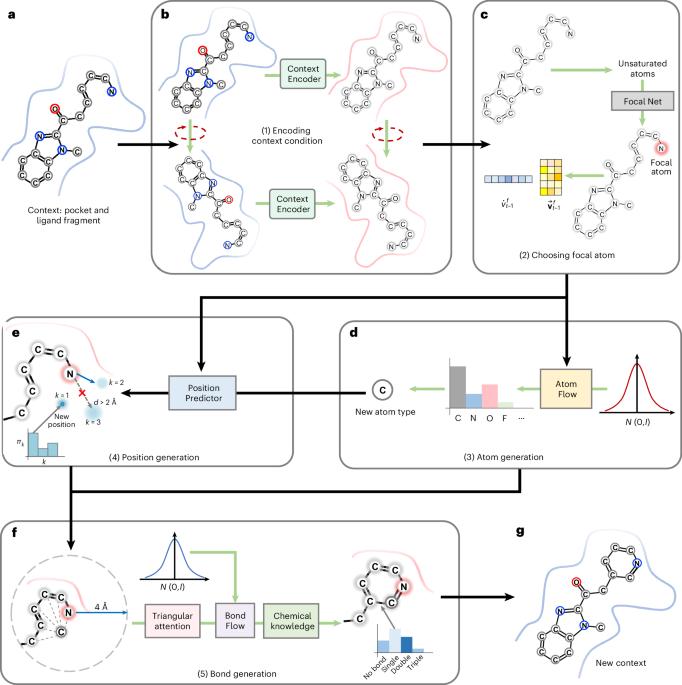
PocketFlow 是一种基于数据和知识驱动的结构分子生成模型
基于深度学习的分子生成在许多领域都有广泛的应用,尤其是药物发现领域。然而,目前大多数深度生成模型都是基于配体的,在分子生成过程中没有考虑化学知识,往往导致成功率相对较低。在此,我们提出了一个基于结构的分子生成框架,其中明确考虑了化学知识(命名为 PocketFlow),该框架可生成蛋白质结合口袋内的新型配体分子。在各种计算评估中,PocketFlow 表现出了最先进的性能,生成的分子 100% 化学有效且高度类药物。消融实验证明了化学知识在确保生成分子的有效性和药物相似性方面的关键作用。我们将 PocketFlow 应用于两个与表观遗传调控相关的新靶蛋白 HAT1 和 YTHDC1,并成功获得了经湿法实验室验证的生物活性化合物。这些活性化合物与目标蛋白的结合模式与分子对接预测的模式接近,并得到了 X 射线晶体结构的进一步证实。所有结果表明,PocketFlow 是一种有用的深度生成模型,能够在给定蛋白质结合口袋的情况下从头生成创新的生物活性分子。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


