Accurate detection of cell deformability tracking in hydrodynamic flow by coupling unsupervised and supervised learning
Abstract
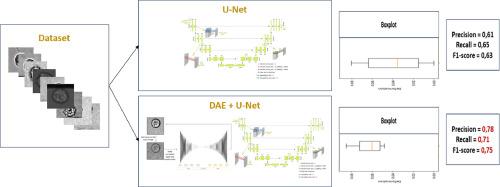
The using of deep learning methods in medical images has been successfully used for various applications, including cell segmentation and deformability detection, thereby contributing significantly to advancements in medical analysis. Cell deformability is a fundamental criterion, which must be measured easily and accurately. One common approach for measuring cell deformability is to use microscopy techniques. Recent works have been efforts to develop more advanced and automated methods for measuring cell deformability based on microscopic images, but cell membrane segmentation techniques are still difficult to achieve with precision because of the quality of images. In this paper, we introduce a novel algorithm for cell segmentation that addresses the challenge of microscopic images. AD-MSC cells were controlled by a microfluidic-based system and cell images were acquired by an ultra-fast camera with variable frequency connected to a powerful computer to collect data. The proposed algorithm has a combination of two main components: denoising images using unsupervised learning and cell segmentation and deformability detection using supervised learning which aim to enhance image quality without the need for expensive materials and expert intervention and segment cell deformability with more precision. The contribution of this paper is the combination of two neural networks that treat the database more easily and without the presence of experts. This approach is used to have faster results with high performance according to low datasets from microscopy even with noisy microscopic images. The precision increases to 81 % when we combine DAE with U-Net, compared to 78 % when adding VAE to U-Net and 59 % when using only U-Net.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


