Anoikis-related genes as potential prognostic biomarkers in gastric cancer: A multilevel integrative analysis and predictive therapeutic value
Abstract
Background
Gastric cancer (GC) is a frequent malignancy of the gastrointestinal tract. Exploring the potential anoikis mechanisms and pathways might facilitate GC research.
Purpose
The authors aim to determine the significance of anoikis-related genes (ARGs) in GC prognosis and explore the regulatory mechanisms in epigenetics.
Methods
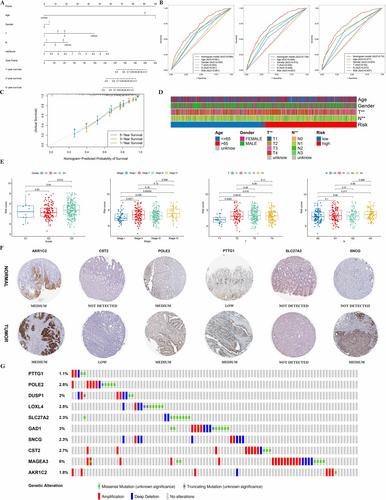
After describing the genetic and transcriptional alterations of ARGs, we searched differentially expressed genes (DEGs) from the cancer genome atlas and gene expression omnibus databases to identify major cancer marker pathways. The non-negative matrix factorisation algorithm, Lasso, and Cox regression analysis were used to construct a risk model, and we validated and assessed the nomogram. Based on multiple levels and online platforms, this research evaluated the regulatory relationship of ARGs with GC.
Results
Overexpression of ARGs is associated with poor prognosis, which modulates immune signalling and promotes anti-anoikis. The consistency of the DEGs clustering with weighted gene co-expression network analysis results and the nomogram containing 10 variable genes improved the clinical applicability of ARGs. In anti-anoikis mode, cytology, histology, and epigenetics could facilitate the analysis of immunophenotypes, tumour immune microenvironment (TIME), and treatment prognosis.
Conclusion
A novel anoikis-related prognostic model for GC is constructed, and the significance of anoikis-related prognostic genes in the TIME and the metabolic pathways of tumours is initially explored.


 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


