Comparison of immune-related gene signatures and immune infiltration features in early- and late-onset preeclampsia
Abstract
Background
Preeclampsia, a severe pregnancy syndrome, is widely accepted divided into early- and late-onset preeclampsia (EOPE and LOPE) based on the onset time of preeclampsia, with distinct pathophysiological origins. However, the molecular mechanism especially immune-related mechanisms for EOPE and LOPE is currently obscure. In the present study, we focused on placental immune alterations between EOPE and LOPE and search for immune-related biomarkers that could potentially serve as potential therapeutic targets through bioinformatic analysis.
Methods
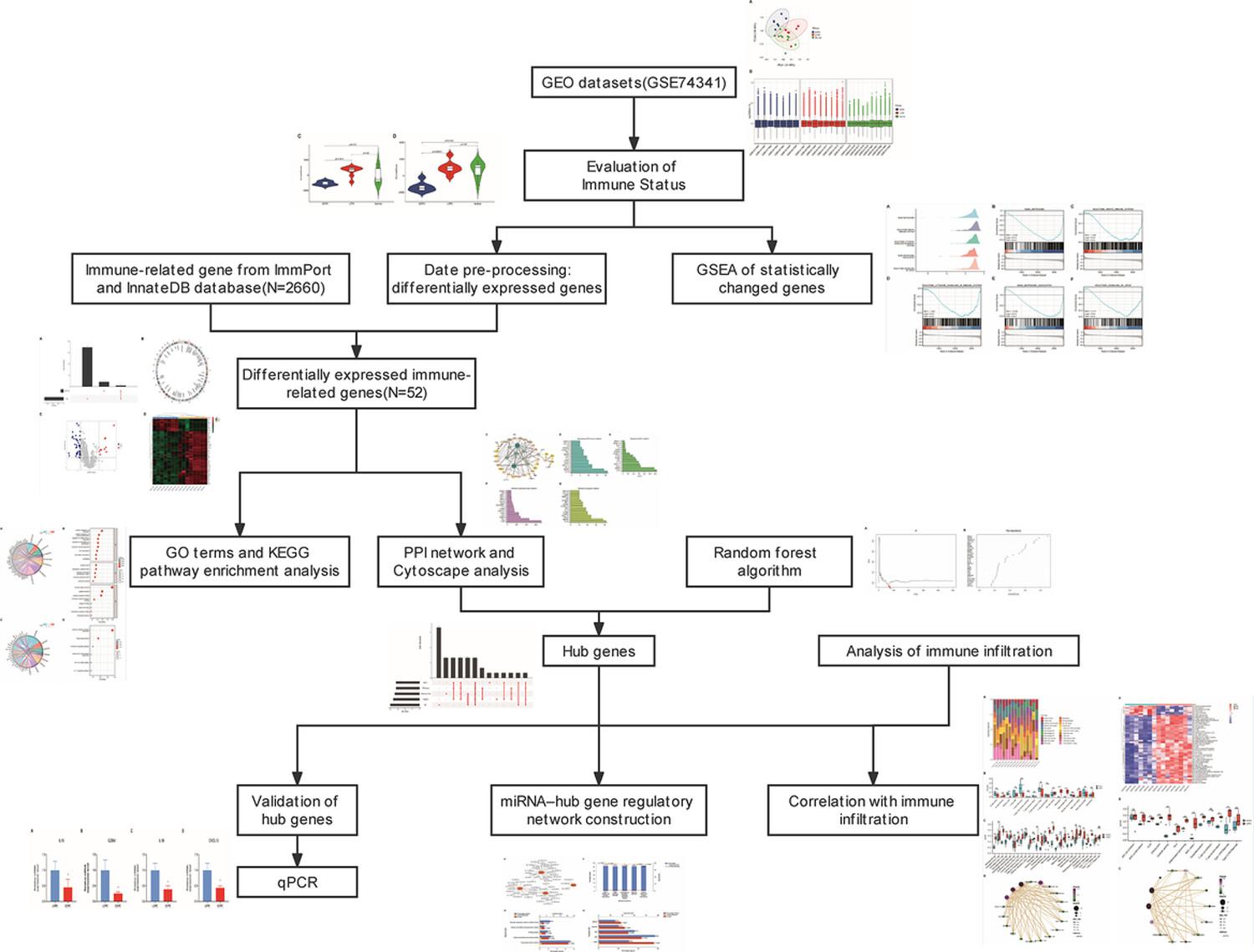
The gene expression profiling data was obtained from the Gene Expression Omnibus database. ESTIMATE algorithm and Gene Set Enrichment Analysis were employed to evaluate the immune status. The intersection of differentially expressed genes in GSE74341 series and immune-related genes set screened differentially expressed immune-related genes. Protein–protein interaction network and random forest were used to identify hub genes with a validation by a quantitative real-time PCR. Kyoto Encyclopedia of Genes and Genomes pathways, Gene Ontology and gene set variation analysis were utilized to conduct biological function and pathway enrichment analyses. Single-sample gene set enrichment analysis and CIBERSORTx tools were employed to calculate the immune cell infiltration score. Correlation analyses were evaluated by Pearson correlation analysis. Hub genes-miRNA network was performed by the NetworkAnalyst online tool.
Results
Immune score and stromal score were all lower in EOPE samples. The immune system-related gene set was significantly downregulated in EOPE compared to LOPE samples. Four hub differentially expressed immune-related genes (IL15, GZMB, IL1B and CXCL12) were identified based on a protein–protein interaction network and random forest. Quantitative real-time polymerase chain reaction validated the lower expression levels of four hub genes in EOPE compared to LOPE samples. Immune cell infiltration analysis found that innate and adaptive immune cells were apparent lacking in EOPE samples compared to LOPE samples. Cytokine-cytokine receptor, para-inflammation, major histocompatibility complex class I and T cell co-stimulation pathways were significantly deficient and highly correlated with hub genes. We constructed a hub genes-miRNA regulatory network, revealing the correlation between hub genes and hsa-miR-374a-5p, hsa-miR-203a-3p, hsa-miR-128-3p, hsa-miR-155-3p, hsa-miR-129-2-3p and hsa-miR-7-5p.
Conclusions
The innate and adaptive immune systems were severely impaired in placentas of EOPE. Four immune-related genes (IL15, GZMB, IL1B and CXCL12) were closely correlated with immune-related pathogenesis of EOPE. The result of our study may provide a new basis for discriminating between EOPE and LOPE and acknowledging the role of the immune landscape in the eventual interference and tailored treatment of EOPE.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


