Sparks of function by de novo protein design
IF 41.7
1区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Information in proteins flows from sequence to structure to function, with each step causally driven by the preceding one. Protein design is founded on inverting this process: specify a desired function, design a structure executing this function, and find a sequence that folds into this structure. This ‘central dogma’ underlies nearly all de novo protein-design efforts. Our ability to accomplish these tasks depends on our understanding of protein folding and function and our ability to capture this understanding in computational methods. In recent years, deep learning-derived approaches for efficient and accurate structure modeling and enrichment of successful designs have enabled progression beyond the design of protein structures and towards the design of functional proteins. We examine these advances in the broader context of classical de novo protein design and consider implications for future challenges to come, including fundamental capabilities such as sequence and structure co-design and conformational control considering flexibility, and functional objectives such as antibody and enzyme design. Chu and colleagues discuss recent developments in de novo protein design.
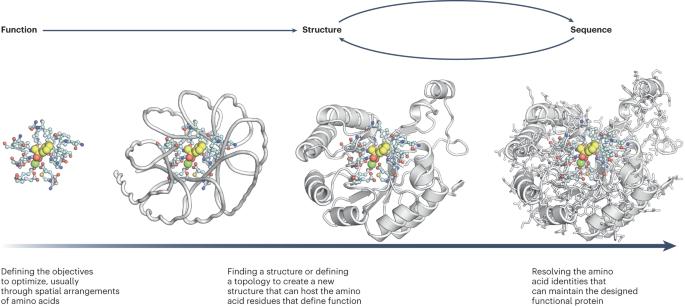
全新蛋白质设计的功能火花。
蛋白质中的信息从序列到结构再到功能,每一步都由前一步因果驱动。蛋白质设计的基础是颠倒这一过程:指定所需的功能,设计执行该功能的结构,并找到折叠到该结构中的序列。这一 "中心教条 "是几乎所有全新蛋白质设计工作的基础。我们完成这些任务的能力取决于我们对蛋白质折叠和功能的理解,以及用计算方法捕捉这种理解的能力。近年来,由深度学习衍生出的高效准确的结构建模方法和成功设计的富集方法使我们得以超越蛋白质结构设计,向功能蛋白质设计迈进。我们将在经典从头蛋白质设计的大背景下审视这些进展,并考虑其对未来挑战的影响,包括序列和结构协同设计、考虑灵活性的构象控制等基本能力,以及抗体和酶设计等功能目标。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature biotechnology
工程技术-生物工程与应用微生物
CiteScore
63.00
自引率
1.70%
发文量
382
审稿时长
3 months
期刊介绍:
Nature Biotechnology is a monthly journal that focuses on the science and business of biotechnology. It covers a wide range of topics including technology/methodology advancements in the biological, biomedical, agricultural, and environmental sciences. The journal also explores the commercial, political, ethical, legal, and societal aspects of this research.
The journal serves researchers by providing peer-reviewed research papers in the field of biotechnology. It also serves the business community by delivering news about research developments. This approach ensures that both the scientific and business communities are well-informed and able to stay up-to-date on the latest advancements and opportunities in the field.
Some key areas of interest in which the journal actively seeks research papers include molecular engineering of nucleic acids and proteins, molecular therapy, large-scale biology, computational biology, regenerative medicine, imaging technology, analytical biotechnology, applied immunology, food and agricultural biotechnology, and environmental biotechnology.
In summary, Nature Biotechnology is a comprehensive journal that covers both the scientific and business aspects of biotechnology. It strives to provide researchers with valuable research papers and news while also delivering important scientific advancements to the business community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


