Adaptive immune receptor repertoire analysis
IF 56
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
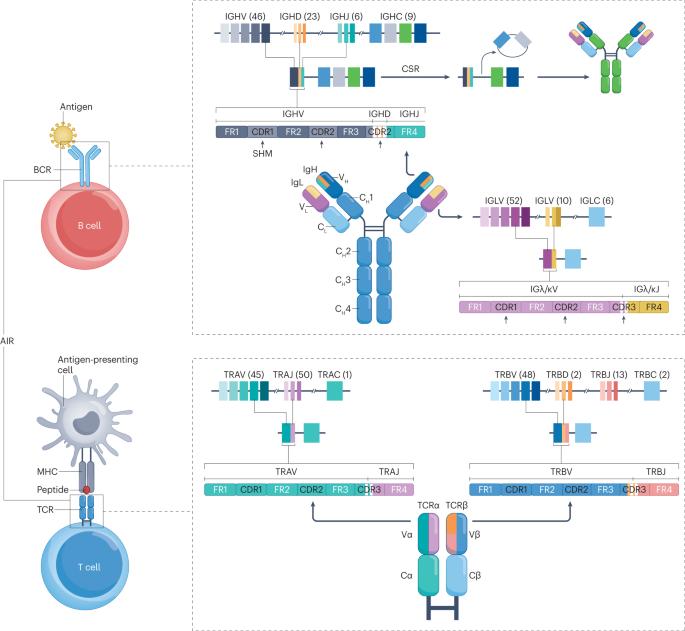
B cell and T cell receptor repertoires compose the adaptive immune receptor repertoire (AIRR) of an individual. The AIRR is a unique collection of antigen-specific receptors that drives adaptive immune responses, which in turn is imprinted in each individual AIRR. This supports the concept that the AIRR could determine disease outcomes, for example in autoimmunity, infectious disease and cancer. AIRR analysis could therefore assist the diagnosis, prognosis and treatment of human diseases towards personalized medicine. High-throughput sequencing, high-dimensional statistical analysis, computational structural biology and machine learning are currently employed to study the shaping and dynamics of the AIRR as a function of time and antigenic challenges. This Primer provides an overview of concepts and state-of-the-art methods that underlie experimental and computational AIRR analysis and illustrates the diversity of relevant applications. The Primer also addresses some of the outstanding challenges in AIRR analysis, such as sampling, sequencing depth, experimental variations and computational biases, while discussing prospects of future AIRR analysis applications for understanding and predicting adaptive immune responses. The adaptive immune receptor repertoire (AIRR) drives adaptive immune responses, which could determine disease outcomes, infectious disease and cancer. Mhanna, Bashour et al. outline the approaches and challenges in AIRR analysis, as well as future developments towards predicting adaptive immune responses.
适应性免疫受体谱系分析
B 细胞和 T 细胞受体谱系组成了个体的适应性免疫受体谱系(AIRR)。AIRR 是驱动适应性免疫反应的抗原特异性受体的独特集合,反过来又烙印在每个个体的 AIRR 中。这支持了 AIRR 可以决定疾病结果的概念,例如在自身免疫、传染病和癌症中。因此,AIRR 分析有助于人类疾病的诊断、预后和治疗,从而实现个性化医疗。目前正在利用高通量测序、高维统计分析、计算结构生物学和机器学习来研究 AIRR 随时间和抗原挑战而变化的形状和动态。本手册概述了实验和计算 AIRR 分析的基本概念和最新方法,并说明了相关应用的多样性。本手册还探讨了 AIRR 分析中的一些突出挑战,如取样、测序深度、实验变化和计算偏差,同时讨论了未来 AIRR 分析在理解和预测适应性免疫反应方面的应用前景。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


