Network pharmacology combined with molecular docking simulations reveal the mechanism of action of Glycyrrhiza for treating pneumonia
IF 1.5
4区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
A well-established mechanism of action for managing pneumonia using Glycyrrhiza is unknown. Using network pharmacology and molecular docking simulations, we investigated the mechanism of action of Glycyrrhiza against pneumonia. To identify the targets of the active components of Glycyrrhiza from the Traditional Chinese Medicine Systems Pharmacology database, oral bioavailability and drug likeness were utilized as indicators. Pneumonia-associated genes were identified and screened from the databases. Integrated analysis was conducted to elucidate the relationship between the active components of Glycyrrhiza and intersecting genes; a comprehensive Glycyrrhiza active component-target gene relationship map was constructed. Intersecting genes underwent Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses to examine their biological functions. A protein–protein interaction network map was constructed to identify hub genes. Molecular docking simulations were performed to investigate binding interactions between hub genes and their corresponding active components. Of the 96 overlapping genes, topological analysis revealed 10 hub genes. Glycyrrhiza exerts therapeutic effects through a multi-target and multipathway approach, suggesting a synergistic treatment for pneumonia. MAPK14 showed a favorable binding affinity with most of the active compounds, indicating that MAPK14 and related compounds in Glycyrrhiza have development potential.
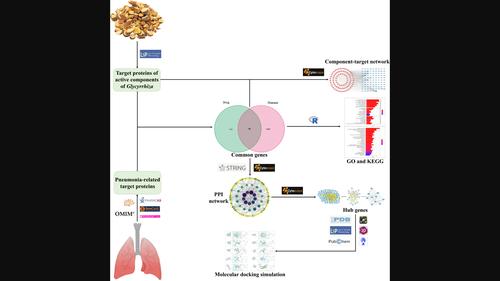
网络药理学结合分子对接模拟揭示甘草治疗肺炎的作用机制
使用甘草治疗肺炎的成熟作用机制尚不清楚。我们利用网络药理学和分子对接模拟研究了甘草治疗肺炎的作用机制。为了从中药系统药理学数据库中确定甘草活性成分的靶点,我们利用口服生物利用度和药物相似度作为指标。从数据库中识别并筛选出肺炎相关基因。通过综合分析,阐明了甘草活性成分与交叉基因之间的关系,构建了甘草活性成分与靶基因的综合关系图谱。对交叉基因进行了基因本体和京都基因组百科全书通路富集分析,以研究其生物学功能。构建了蛋白质-蛋白质相互作用网络图,以确定枢纽基因。进行了分子对接模拟,以研究中心基因与其相应活性成分之间的结合相互作用。在96个重叠基因中,拓扑分析发现了10个中心基因。甘草通过多靶点和多途径的方法发挥治疗作用,这表明甘草对肺炎有协同治疗作用。MAPK14与大多数活性化合物显示出良好的结合亲和力,表明甘草中的MAPK14和相关化合物具有开发潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Peptide Science
Biochemistry, Genetics and Molecular Biology-Biophysics
CiteScore
5.20
自引率
4.20%
发文量
36
期刊介绍:
The aim of Peptide Science is to publish significant original research papers and up-to-date reviews covering the entire field of peptide research. Peptide Science provides a forum for papers exploring all aspects of peptide synthesis, materials, structure and bioactivity, including the use of peptides in exploring protein functions and protein-protein interactions. By incorporating both experimental and theoretical studies across the whole spectrum of peptide science, the journal serves the interdisciplinary biochemical, biomaterials, biophysical and biomedical research communities.
Peptide Science is the official journal of the American Peptide Society.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


