Privacy-preserving record linkage across disparate institutions and datasets to enable a learning health system: The national COVID cohort collaborative (N3C) experience
Abstract
Introduction
Research driven by real-world clinical data is increasingly vital to enabling learning health systems, but integrating such data from across disparate health systems is challenging. As part of the NCATS National COVID Cohort Collaborative (N3C), the N3C Data Enclave was established as a centralized repository of deidentified and harmonized COVID-19 patient data from institutions across the US. However, making this data most useful for research requires linking it with information such as mortality data, images, and viral variants. The objective of this project was to establish privacy-preserving record linkage (PPRL) methods to ensure that patient-level EHR data remains secure and private when governance-approved linkages with other datasets occur.
Methods
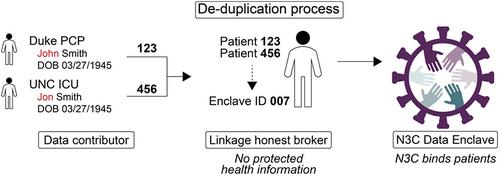
Separate agreements and approval processes govern N3C data contribution and data access. The Linkage Honest Broker (LHB), an independent neutral party (the Regenstrief Institute), ensures data linkages are robust and secure by adding an extra layer of separation between protected health information and clinical data. The LHB's PPRL methods (including algorithms, processes, and governance) match patient records using “deidentified tokens,” which are hashed combinations of identifier fields that define a match across data repositories without using patients' clear-text identifiers.
Results
These methods enable three linkage functions: Deduplication, Linking Multiple Datasets, and Cohort Discovery. To date, two external repositories have been cross-linked. As of March 1, 2023, 43 sites have signed the LHB Agreement; 35 sites have sent tokens generated for 9 528 998 patients. In this initial cohort, the LHB identified 135 037 matches and 68 596 duplicates.
Conclusion
This large-scale linkage study using deidentified datasets of varying characteristics established secure methods for protecting the privacy of N3C patient data when linked for research purposes. This technology has potential for use with registries for other diseases and conditions.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


