Prime editing using CRISPR-Cas12a and circular RNAs in human cells
IF 41.7
1区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Genome editing with prime editors based on CRISPR-Cas9 is limited by the large size of the system and the requirement for a G/C-rich protospacer-adjacent motif (PAM) sequence. Here, we use the smaller Cas12a protein to develop four circular RNA-mediated prime editor (CPE) systems: nickase-dependent CPE (niCPE), nuclease-dependent CPE (nuCPE), split nickase-dependent CPE (sniCPE) and split nuclease-dependent CPE (snuCPE). CPE systems preferentially recognize T-rich genomic regions and possess a potential multiplexing capacity in comparison to corresponding Cas9-based systems. The efficiencies of the nuclease-based systems are up to 10.42%, whereas niCPE and sniCPE reach editing frequencies of up to 24.89% and 40.75% without positive selection in human cells, respectively. A derivative system, called one-sniCPE, combines all three RNA editing components under a single promoter. By arraying CRISPR RNAs for different targets in one circular RNA, we also demonstrate low-efficiency editing of up to four genes simultaneously with the nickase prime editors niCPE and sniCPE. Prime editing with CRISPR-Cas12a broadens targeting scope and enables multiplexing.
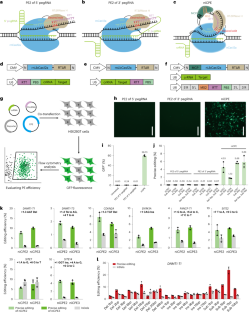

在人体细胞中使用 CRISPR-Cas12a 和环状 RNA 进行基因编辑。
基于CRISPR-Cas9的主编辑器的基因组编辑受到了系统体积大和需要富含G/C的原间隔相邻基序(PAM)的限制。在这里,我们利用较小的 Cas12a 蛋白开发了四种环形 RNA 介导的质粒编辑器(CPE)系统:缺口酶依赖 CPE(niCPE)、核酸酶依赖 CPE(nuCPE)、分裂缺口酶依赖 CPE(sniCPE)和分裂核酸酶依赖 CPE(snuCPE)。与相应的基于 Cas9 的系统相比,CPE 系统能优先识别富含 T 的基因组区域,并具有潜在的复用能力。基于核酸酶的系统的效率高达 10.42%,而 niCPE 和 sniCPE 在人类细胞中的编辑频率分别高达 24.89% 和 40.75%,且无阳性选择。一种名为 one-sniCPE 的衍生系统将所有三种 RNA 编辑元件结合在一个启动子下。通过在一个环形 RNA 中排列针对不同靶点的 CRISPR RNA,我们还证明了使用缺口酶素编辑器 niCPE 和 sniCPE 可同时低效编辑多达四个基因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature biotechnology
工程技术-生物工程与应用微生物
CiteScore
63.00
自引率
1.70%
发文量
382
审稿时长
3 months
期刊介绍:
Nature Biotechnology is a monthly journal that focuses on the science and business of biotechnology. It covers a wide range of topics including technology/methodology advancements in the biological, biomedical, agricultural, and environmental sciences. The journal also explores the commercial, political, ethical, legal, and societal aspects of this research.
The journal serves researchers by providing peer-reviewed research papers in the field of biotechnology. It also serves the business community by delivering news about research developments. This approach ensures that both the scientific and business communities are well-informed and able to stay up-to-date on the latest advancements and opportunities in the field.
Some key areas of interest in which the journal actively seeks research papers include molecular engineering of nucleic acids and proteins, molecular therapy, large-scale biology, computational biology, regenerative medicine, imaging technology, analytical biotechnology, applied immunology, food and agricultural biotechnology, and environmental biotechnology.
In summary, Nature Biotechnology is a comprehensive journal that covers both the scientific and business aspects of biotechnology. It strives to provide researchers with valuable research papers and news while also delivering important scientific advancements to the business community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


