SSR1 and CKAP4 as potential biomarkers for intervertebral disc degeneration based on integrated bioinformatics analysis
Abstract
Background
Intervertebral disc degeneration (IDD) is a significant cause of low back pain and poses a significant public health concern. Genetic factors play a crucial role in IDD, highlighting the need for a better understanding of the underlying mechanisms.
Aim
The aim of this study was to identify potential IDD-related biomarkers using a comprehensive bioinformatics approach and validate them in vitro.
Materials and Methods
In this study, we employed several analytical approaches to identify the key genes involved in IDD. We utilized weighted gene coexpression network analysis (WGCNA), MCODE, LASSO algorithms, and ROC curves to identify the key genes. Additionally, immune infiltrating analysis and a single-cell sequencing dataset were utilized to further explore the characteristics of the key genes. Finally, we conducted in vitro experiments on human disc tissues to validate the significance of these key genes in IDD.
Results
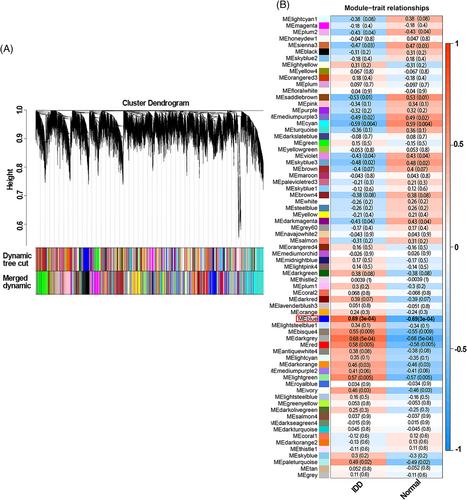
we obtained gene expression profiles from the GEO database (GSE23130 and GSE15227) and identified 1015 DEGs associated with IDD. Using WGCNA, we identified the blue module as significantly related to IDD. Among the DEGs, we identified 47 hub genes that overlapped with the genes in the blue module, based on criteria of |logFC| ≥ 2.0 and p.adj <0.05. Further analysis using both MCODE and LASSO algorithms enabled us to identify five key genes, of which CKAP4 and SSR1 were validated by GSE70362, demonstrating significant diagnostic value for IDD. Additionally, immune infiltrating analysis revealed that monocytes were significantly correlated with the two key genes. We also analyzed a single-cell sequencing dataset, GSE199866, which showed that both CKAP4 and SSR1 were highly expressed in fibrocartilage chondrocytes. Finally, we validated our findings in vitro by performing real time polymerase chain reaction (RT-PCR) and immunohistochemistry (IHC) on 30 human disc samples. Our results showed that CKAP4 and SSR1 were upregulated in degenerated disc samples. Taken together, our findings suggest that CKAP4 and SSR1 have the potential to serve as disease biomarkers for IDD.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


