Alternative splicing and environmental adaptation in wild house mice
IF 3.1
2区 生物学
Q2 ECOLOGY
引用次数: 0
Abstract
A major goal of evolutionary genetics is to understand the genetic and molecular mechanisms underlying adaptation. Previous work has established that changes in gene regulation may contribute to adaptive evolution, but most studies have focused on mRNA abundance and only a few studies have investigated the role of post-transcriptional processing. Here, we use a combination of exome sequences and short-read RNA-Seq data from wild house mice (Mus musculus domesticus) collected along a latitudinal transect in eastern North America to identify candidate genes for local adaptation through alternative splicing. First, we identified alternatively spliced transcripts that differ in frequency between mice from the northern-most and southern-most populations in this transect. We then identified the subset of these transcripts that exhibit clinal patterns of variation among all populations in the transect. Finally, we conducted association studies to identify cis-acting splicing quantitative trait loci (cis-sQTL), and we identified cis-sQTL that overlapped with previously ascertained targets of selection from genome scans. Together, these analyses identified a small set of alternatively spliced transcripts that may underlie environmental adaptation in house mice. Many of these genes have known phenotypes associated with body size, a trait that varies clinally in these populations. We observed no overlap between these genes and genes previously identified by changes in mRNA abundance, indicating that alternative splicing and changes in mRNA abundance may provide separate molecular mechanisms of adaptation.
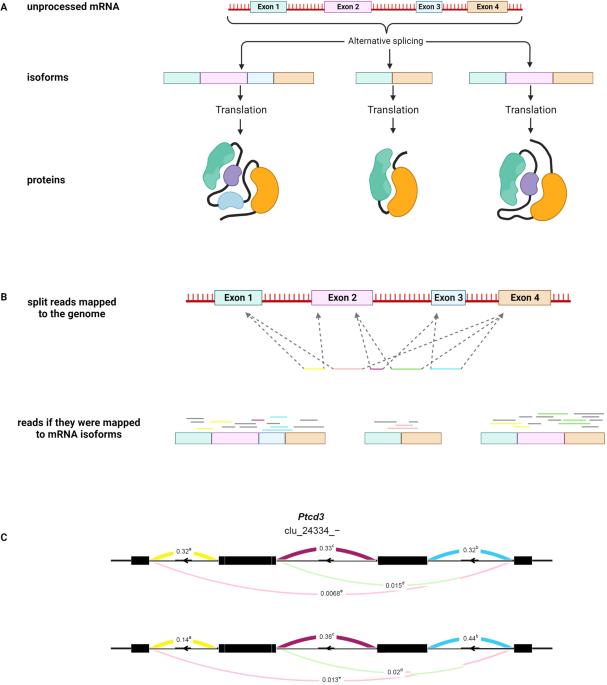
野生家鼠的选择性剪接与环境适应。
进化遗传学的一个主要目标是了解适应的遗传和分子机制。先前的研究已经确定基因调控的变化可能有助于适应性进化,但大多数研究都集中在mRNA丰度上,只有少数研究调查了转录后加工的作用。在这里,我们结合了外显子组序列和短读RNA-Seq数据,这些数据来自北美东部沿纬度样带收集的野生家鼠(Mus musculus domesticus),通过选择性剪接确定了局部适应的候选基因。首先,我们确定了该样带中最北和最南种群小鼠之间频率不同的选择性剪接转录本。然后,我们确定了这些转录本的子集,这些转录本在样带的所有种群中表现出临床模式的变化。最后,我们进行了关联研究,以确定顺式作用剪接数量性状位点(cis-sQTL),并从基因组扫描中确定了与先前确定的选择靶点重叠的顺式sqtl。总之,这些分析确定了一小组可选择剪接的转录本,可能是家鼠环境适应的基础。这些基因中的许多都有已知的与体型相关的表型,而体型在这些人群中是不同的。我们观察到这些基因与之前通过mRNA丰度变化确定的基因之间没有重叠,这表明选择性剪接和mRNA丰度的变化可能提供了不同的分子适应机制。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Heredity
生物-进化生物学
CiteScore
7.50
自引率
2.60%
发文量
84
审稿时长
4-8 weeks
期刊介绍:
Heredity is the official journal of the Genetics Society. It covers a broad range of topics within the field of genetics and therefore papers must address conceptual or applied issues of interest to the journal''s wide readership
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


