Homologous recombination deficiency reflects the heterogeneity and monitoring treatment response for patients with breast cancer
Abstract
Background
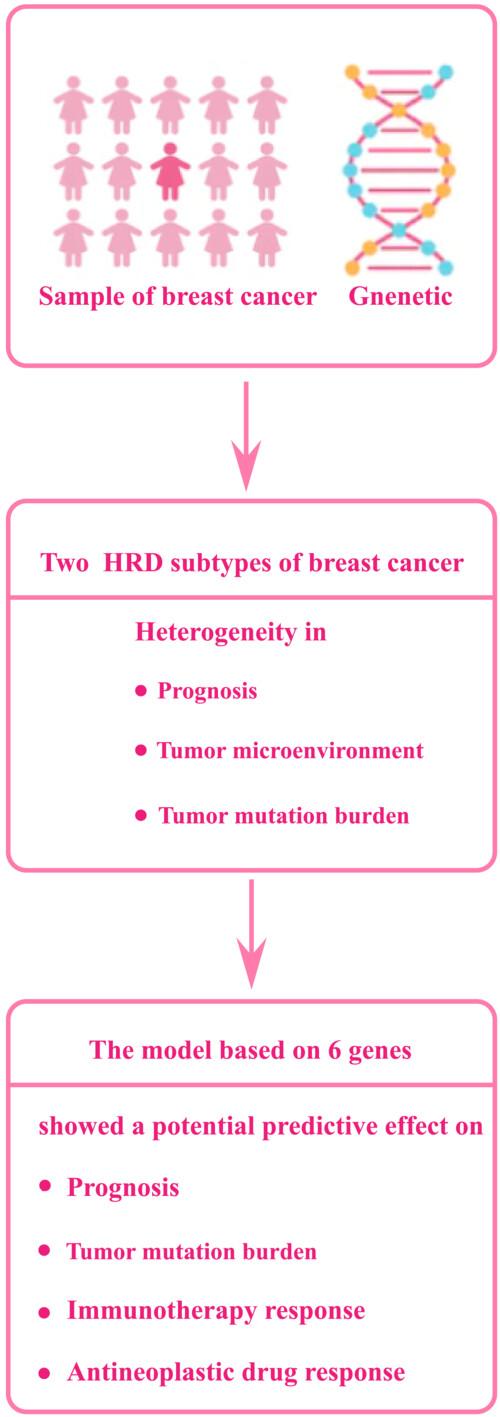
In breast cancer (BC), homologous recombination defect (HRD) is a common carcinogenic mechanism. It is meaningful to classify BC according to HRD biomarkers and to develop a platform for identifying BC molecular features, pathological features and therapeutic responses.
Methods
In total, 109 HRD genes were collected and screened by univariate Cox regression analysis to determine the prognostic genes, which were used to construct a consensus matrix to identify BC subtype. Differentially expressed genes (DEGs) were filtered by the Limma package and screened by random forest analysis to build a model to analyze the immunotherapy response and sensitivity and prognosis of patients suffering from BC to different drugs.
Results
Thirteen out of 109 HRD genes were prognostic genes of BC, and BC was classified into two subgroups based on their expression. Cluster 1 had a significantly backward survival outcome and a significantly higher adaptive immunity score relative to cluster 2. Six genes were identified by random forest analysis as factors for developing the model. The model provided a prediction called risk score, which showed a significant stratification effect on BC prognosis, immunotherapy response and IC50 values of 62 drugs.
Conclusions
In the present study, two HRD subtypes of BC were successfully identified, for which mutation and immunological features were determined. A model based on differential genes of HRD subtypes was established, which was a potential predictor of prognosis, immunotherapy response and drug sensitivity of BC.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


