Genomic analysis of geographical structure and diversity in the capercaillie (Tetrao urogallus)
IF 1.7
3区 环境科学与生态学
Q2 BIODIVERSITY CONSERVATION
引用次数: 0
Abstract
Abstract The capercaillie is widespread throughout the boreal forests of northern Eurasia, but faces serious conservation challenges in the southernmost mountain ranges, where the populations are fragmented and some are critically endangered. To develop effective conservation strategies for these populations, it is essential to have information on both their genetic diversity and the genetic structure of the species. In this work, we used a reduced representation (ddRAD) genomic sequencing technique to analyze the genetic structure of the capercaillie across its European range and to assess the inbreeding levels in some of the most threatened populations. Our population structure analysis suggested the existence of two evolutionarily significant units, one formed by the two populations from the Iberian Peninsula and the other by the populations from the rest of Europe. Genetic diversity showed a significant decrease in the Iberian populations with respect to the other European populations. An isolation-with-migration model supported these results and allowed us to estimate the parameters of the population tree. The inbreeding coefficients estimated for the Iberian capercaillies showed relatively low levels in the Pyrenees. However, some individuals with very high inbreeding values were detected in the Cantabrian Mountains, suggesting that some of its subpopulations are substantially isolated. The population structure results and the genomic monitoring method we used to assess inbreeding levels may be crucial for the conservation and recovery of the most endangered capercaillie populations.
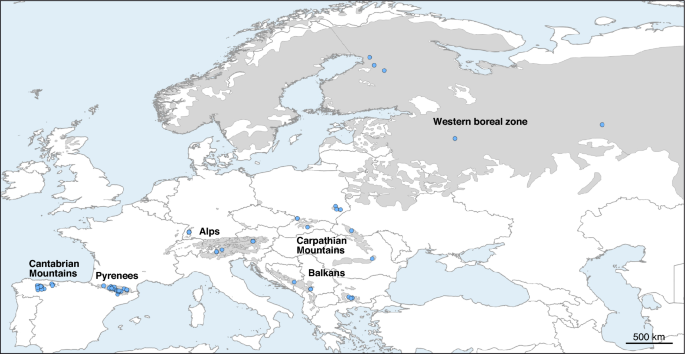
毛猴属(Tetrao urogallus)地理结构和多样性的基因组分析
摘要/ Abstract摘要:狐猴广泛分布于欧亚大陆北部的北方森林中,但在最南端的山脉地区面临着严重的保护挑战,种群分散,有些濒临灭绝。为了对这些种群制定有效的保护策略,掌握其遗传多样性和物种遗传结构的信息是必不可少的。在这项工作中,我们使用了减少表征(ddRAD)基因组测序技术来分析整个欧洲范围的capercaillie的遗传结构,并评估了一些最受威胁的种群的近交水平。我们的种群结构分析表明存在两个进化上重要的单位,一个由来自伊比利亚半岛的两个种群组成,另一个由来自欧洲其他地区的种群组成。遗传多样性表明,伊比利亚人口与其他欧洲人口相比显著减少。隔离-迁移模型支持这些结果,并使我们能够估计种群树的参数。伊比利亚的近交系数估计在比利牛斯山脉相对较低。然而,在坎塔布连山脉中发现了一些近交价值非常高的个体,这表明它的一些亚种群基本上是孤立的。种群结构结果和近交水平的基因组监测方法对保护和恢复最濒危的毛猴种群具有重要意义。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Conservation Genetics
环境科学-生物多样性保护
CiteScore
3.80
自引率
4.50%
发文量
58
审稿时长
1 months
期刊介绍:
Conservation Genetics promotes the conservation of biodiversity by providing a forum for data and ideas, aiding the further development of this area of study. Contributions include work from the disciplines of population genetics, molecular ecology, molecular biology, evolutionary biology, systematics, forensics, and others. The focus is on genetic and evolutionary applications to problems of conservation, reflecting the diversity of concerns relevant to conservation biology. Studies are based on up-to-date technologies, including genomic methodologies. The journal publishes original research papers, short communications, review papers and perspectives.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


