Autophagy dark genes: Can we find them with machine learning?
IF 3.1
Q2 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
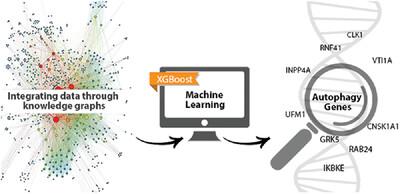
Identifying novel autophagy (ATG) associated genes in humans remains an important task for understanding this fundamental physiological process. Machine learning (ML) can highlight potentially “missing pieces” linking core ATG genes with understudied, “dark” genes by mining functional genomic data. Here, a set of 103 (out of 288 genes from the Autophagy Database) was used as training set, based on ATG-associated terms annotated from 3 secondary sources: GO (gene ontology), Kyoto Encyclopedia of Genes and Genomes pathway, and UniProt keywords, as additional confirmation of their importance in ATG. As negative labels, an OMIM list of genes associated with monogenic diseases was used (after excluding the 288 ATG-associated genes). Data related to these genes from 17 different sources were compiled and used to derive a trained MetaPath/XGBoost (MPxgb) ML model for distinguishing ATG and non-ATG genes (10-fold cross-validated, 100-times randomized models, median area under the curve = 0.994 ± 0.008). Sixteen ATG-relevant variables explained 64% of the total model gain. Overall, 23% of the top 251 predicted genes are annotated in the Autophagy Database, whereas 193 genes (77%) are not. In 2019, we suggested that some of these 193 genes may represent “ATG dark genes.” A literature search in 2022 for those top 20 predicted ATG dark genes found that 9 were subsequently reported as ATG genes during the intervening 3.5 years. A post-factum evaluation of data leakage (the presence of ATG-associated terms in the top 40 ML features) confirms that 7 out of these 9 genes and 2 out of 3 other recently validated predictions from the bottom 20 are novel. Those genes with the largest number of ATG features would be most likely to yield valuable experimental insights. Modern high-throughput testing would be capable of spanning the full 193 ATG genes list reported here. Our analysis demonstrates that ML can guide genomics research to gain a more complete functional and pathway annotation of complex processes. Key points – A knowledge-graph based machine learning model was designed for predicting unknown autophagy genes via mining functional genomic data. – Literature search validated predicted genes. – Our machine learning models could be generalized and applied to other genomic libraries to uncover dark genes for various functions.
自噬暗基因:我们能用机器学习找到它们吗?
在人类中鉴定新的自噬(ATG)相关基因仍然是理解这一基本生理过程的重要任务。机器学习(ML)可以通过挖掘功能基因组数据,突出连接核心ATG基因与未被充分研究的“暗”基因的潜在“缺失片段”。本文使用自噬数据库288个基因中的103个基因作为训练集,基于3个次要来源注解的ATG相关术语:GO(基因本体)、京都基因与基因组百科全书路径和UniProt关键词,以进一步确认它们在ATG中的重要性。作为阴性标记,使用与单基因疾病相关的OMIM基因列表(在排除288个atg相关基因后)。将来自17个不同来源的相关基因数据进行编译,并使用训练后的MetaPath/XGBoost (MPxgb) ML模型来区分ATG和非ATG基因(10倍交叉验证,100倍随机化模型,曲线下中位数面积= 0.994±0.008)。16个atg相关变量解释了64%的模型总增益。总的来说,在自噬数据库中,前251个预测基因中有23%被注释,而193个基因(77%)没有。2019年,我们提出这193个基因中的一些可能代表“ATG暗基因”。2022年对前20个预测ATG暗基因的文献检索发现,在这中间的3.5年里,有9个被报道为ATG基因。对数据泄露的事后评估(前40个ML特征中与atg相关的术语的存在)证实,这9个基因中的7个以及最近验证的后20个预测中的3个中的2个是新的。那些具有最多ATG特征的基因最有可能产生有价值的实验见解。现代高通量检测将能够跨越完整的193个ATG基因列表。我们的分析表明,机器学习可以指导基因组学研究,以获得更完整的复杂过程的功能和途径注释。设计了一个基于知识图谱的机器学习模型,通过挖掘功能基因组数据来预测未知的自噬基因。-文献检索验证了预测的基因。-我们的机器学习模型可以推广并应用于其他基因组文库,以发现各种功能的暗基因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


