Prevalent use and evolution of exonic regulatory sequences in the human genome
IF 3.1
Q2 MULTIDISCIPLINARY SCIENCES
引用次数: 0
Abstract
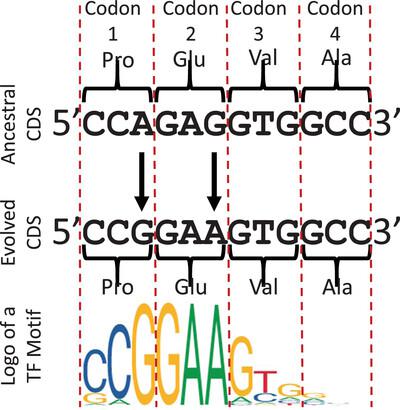
Abstract It has long been known that exons can serve as cis‐ regulatory sequences, such as enhancers. However, the prevalence of such dual‐use of exons and how they evolve remain elusive. Based on our recently predicted, highly accurate large sets of cis ‐regulatory module candidates (CRMCs) and non‐CRMCs in the human genome, we find that exonic transcription factor binding sites (TFBSs) occupy at least a third of the total exon lengths, and 96.7% of genes have exonic TFBSs. Both A/T and C/G in exonic TFBSs are more likely under evolutionary constraints than those in non‐CRMC exons. Exonic TFBSs in codons tend to encode loops rather than more critical helices and strands in protein structures, while exonic TFBSs in untranslated regions (UTRs) tend to avoid positions where known UTR‐related functions are located. Moreover, active exonic TFBSs tend to be in close physical proximity to distal promoters whose genes have elevated transcription levels. These results suggest that exonic TFBSs might be more prevalent than originally thought and likely in dual‐use. We proposed a parsimonious model that well explains the observed evolutionary behaviors of exonic TFBS as well as how a stretch of codons evolve into a TFBS. Key points There are more exonic regulatory sequences in the human genome than originally thought. Exonic transcription factor binding sites are more likely under negative selection or positive selection than counterpart nonregulatory sequences. Exonic transcription factor binding sites tend to be located in genome sequences that encode less critical loops in protein structures, or in less critical parts in 5′ and 3′ untranslated regions.
外显子调控序列在人类基因组中的普遍使用和进化
人们早就知道外显子可以作为顺式调控序列,如增强子。然而,这种双重使用外显子的普遍性及其如何进化仍然是难以捉摸的。基于我们最近预测的、高度精确的人类基因组中顺式调控模块候选(crmc)和非crmc的大集合,我们发现外显子转录因子结合位点(TFBSs)至少占据了总外显子长度的三分之一,96.7%的基因具有外显子TFBSs。外显子TFBSs中的A/T和C/G比非CRMC外显子中的A/T和C/G更可能受到进化限制。密码子中的外显子TFBSs倾向于编码环,而不是蛋白质结构中更关键的螺旋和链,而非翻译区(UTR)中的外显子TFBSs倾向于避开已知UTR相关功能所在的位置。此外,活跃的外显子TFBSs往往在物理上接近远端启动子,其基因的转录水平升高。这些结果表明,外显子TFBSs可能比原先认为的更为普遍,并且可能具有双重用途。我们提出了一个简洁的模型,很好地解释了观察到的外显子TFBS的进化行为,以及一段密码子如何进化成TFBS。人类基因组中的外显子调控序列比原先认为的要多。外显子转录因子结合位点在负选择或正选择下比对应的非调控序列更容易出现。外显子转录因子结合位点往往位于编码蛋白质结构中不太关键环的基因组序列中,或者位于5 '和3 '非翻译区中不太关键的部分。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


