Pleistocene-dated genomic divergence of avocado trees supports cryptic diversity in the Colombian germplasm
IF 1.6
3区 生物学
Q2 FORESTRY
引用次数: 0
Abstract
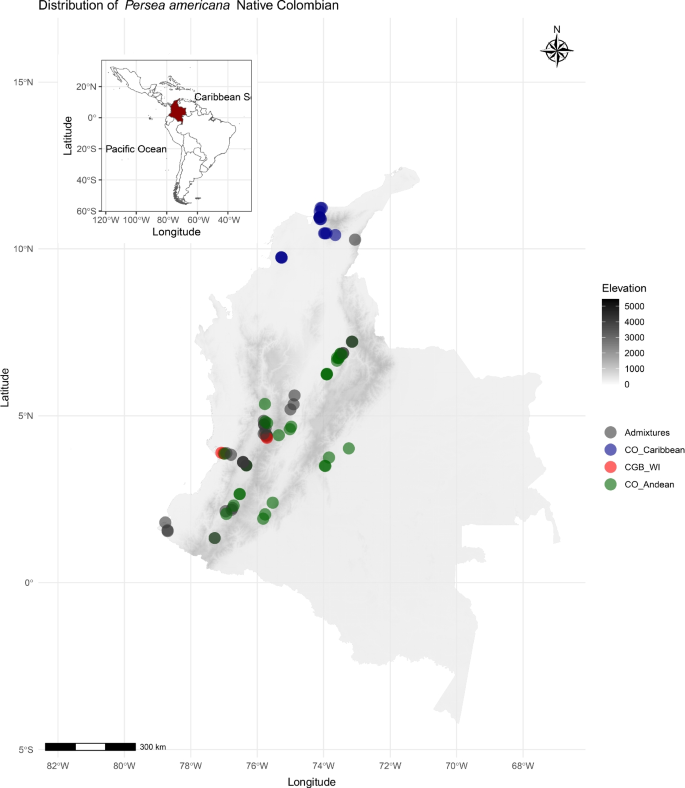
Abstract Genomic characterization of ex situ plant collections optimizes the utilization of genetic resources by identifying redundancies among accessions, capturing cryptic variation, establishing reference collections, and ultimately assisting pre-breeding and breeding efforts. Yet, the integration of evolutionary genomic analyses is often lacking when studying the biodiversity of crop gene pools. Such is the case in the avocado, Persea americana Mill., an iconic American fruit tree crop that has seen an unprecedented expansion worldwide because of its nutritional properties. However, given a very restricted number of commercial clones, avocado plantations are becoming more vulnerable to diseases and climate change. Therefore, exploring new sources of evolutionary novelty and genetic diversity beyond the commercial varieties derived from traditional genetic pools in Mexico and Central America is imperative. To fill this gap, we aimed to characterize the genomic diversity of Colombian avocado trees. Specifically, we constructed reduced representation genomic libraries to genotype by sequencing 144 accessions from the Colombian National genebank and 240 materials from local commercial orchards in the Colombian northwest Andes. We merged the resulting reads with available sequences of reference genotypes from known avocado groups (also named as races), Mexican, Guatemalan, and West Indian, to discover 4931 SNPs. We then analyzed the population structure and phylogenetic diversity, and reconstructed evolutionary scenarios, possibly leading to new genetic groups in Colombian germplasm. We detected demographic stratification despite evidence of intergroup gene flow. Besides the classical three avocado groups, we found an exclusive Colombian group with a possible genetic substructure related to the geographical origin (Andean and Caribbean). Phylogenetic and ABC demographic modeling suggested that the Colombian group evolved in the Pleistocene before human agriculture started, and its closest relative from the three recognized races would be the West Indian group. We conclude that northwest South America offers a cryptic source of allelic novelty capable of boosting avocado pre-breeding strategies to select rootstock candidates well adapted to specific eco-geographical regions in Colombia and abroad.
更新世时代的鳄梨树基因组分化支持哥伦比亚种质的隐性多样性
迁地植物的基因组鉴定可以通过鉴定遗传资源的冗余性、捕获隐性变异、建立参考资料、最终辅助育种和育种工作来优化遗传资源的利用。然而,在研究作物基因库的生物多样性时,往往缺乏进化基因组分析的整合。这就是鳄梨Persea americana Mill的情况。这是一种标志性的美国果树作物,由于其营养特性,在全球范围内得到了前所未有的扩张。然而,由于商业克隆数量非常有限,牛油果种植园越来越容易受到疾病和气候变化的影响。因此,在墨西哥和中美洲传统基因库衍生的商业品种之外,探索进化新颖性和遗传多样性的新来源是势在必行的。为了填补这一空白,我们的目标是表征哥伦比亚鳄梨树的基因组多样性。具体来说,我们通过对来自哥伦比亚国家基因库的144份材料和来自哥伦比亚西北部安第斯山脉当地商业果园的240份材料进行测序,构建了减少代表性的基因组文库。我们将结果读取与已知鳄梨群体(也称为种族),墨西哥,危地马拉和西印度的可用参考基因型序列合并,发现4931个snp。然后,我们分析了种群结构和系统发育多样性,并重建了进化情景,可能导致哥伦比亚种质资源中新的遗传群。尽管存在群体间基因流动的证据,我们还是发现了人口分层。除了经典的三个鳄梨类群外,我们还发现了一个独特的哥伦比亚类群,其遗传亚结构可能与地理起源(安第斯和加勒比)有关。系统发育和ABC人口统计学模型表明,哥伦比亚人在更新世进化,早于人类农业的开始,在三个已知的种族中,与他们最近的亲戚是西印度人。我们得出结论,南美洲西北部提供了一个神秘的等位基因新颖性来源,能够促进鳄梨的预育种策略,以选择适合哥伦比亚和国外特定生态地理区域的候选砧木。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Tree Genetics & Genomes
生物-林学
CiteScore
4.40
自引率
4.20%
发文量
38
审稿时长
2 months
期刊介绍:
Tree Genetics and Genomes is an international, peer-reviewed journal, which provides for the rapid publication of high quality papers covering the areas of forest and horticultural tree genetics and genomics.
Topics covered in this journal include:
Structural, functional and comparative genomics
Evolutionary, population and quantitative genetics
Ecological and physiological genetics
Molecular, cellular and developmental genetics
Conservation and restoration genetics
Breeding and germplasm development
Bioinformatics and databases
Tree Genetics and Genomes publishes four types of papers:
(1) Original Paper
(2) Review
(3) Opinion Paper
(4) Short Communication.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


