{"title":"\"SIT\" with Emx1-NuTRAP Mice: Simultaneous INTACT and TRAP for Paired Transcriptomic and Epigenetic Sequencing.","authors":"Anthony M Raus, Nellie E Nelson, Tyson D Fuller, Autumn S Ivy","doi":"10.1002/cpz1.570","DOIUrl":null,"url":null,"abstract":"<p><p>Epigenetic regulation of transcription is gaining increasing importance in the study of neurobiology. The advent of sequencing technology has enabled the study of this regulation across the entire genome and transcriptome. However, modern methods that allow the correlation of transcriptomic data with epigenomic regulation have had several key limitations, including use of separate tissue sources and detection of low-expression genes. This article describes a method combining isolation of nuclei tagged in specific cell types (INTACT) with translating ribosome affinity purification (TRAP) in the same cell homogenate, referred to as Simultaneous INTACT and TRAP (SIT). We used this technical approach to directly couple transcriptomic sequencing with epigenomic data in neurons derived from the mouse hippocampus. We demonstrate this method with an Emx1-NuTRAP transgenic mouse model. Here, we present protocols for SIT and for the generation and validation of the Emx1-NuTRAP mouse model that we used to demonstrate SIT. These methods enable cell type-specific comparison of translating mRNA and chromatin data from the same set of cells. Using SIT and the Emx1-NuTRAP transgenic mouse model, researchers can compare epigenomic data to transcriptomic data in the same set of hippocampal excitatory neurons. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC. Basic Protocol 1: Emx1-NuTRAP transgenic mouse line for labeling excitatory neurons in the hippocampus Basic Protocol 2: SIT: Simultaneous Isolation of nuclei tagged in specific cell types (INTACT) and Translating ribosome affinity purification (TRAP).</p>","PeriodicalId":11174,"journal":{"name":"Current Protocols","volume":"2 10","pages":"e570"},"PeriodicalIF":0.0000,"publicationDate":"2022-10-01","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9614570/pdf/","citationCount":"1","resultStr":null,"platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current Protocols","FirstCategoryId":"1085","ListUrlMain":"https://doi.org/10.1002/cpz1.570","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"","JCRName":"","Score":null,"Total":0}
引用次数: 1
Abstract
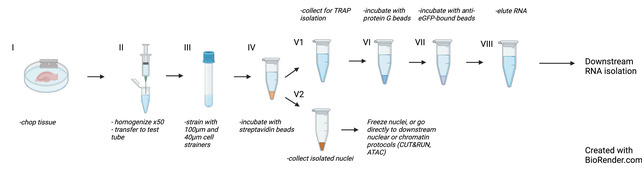
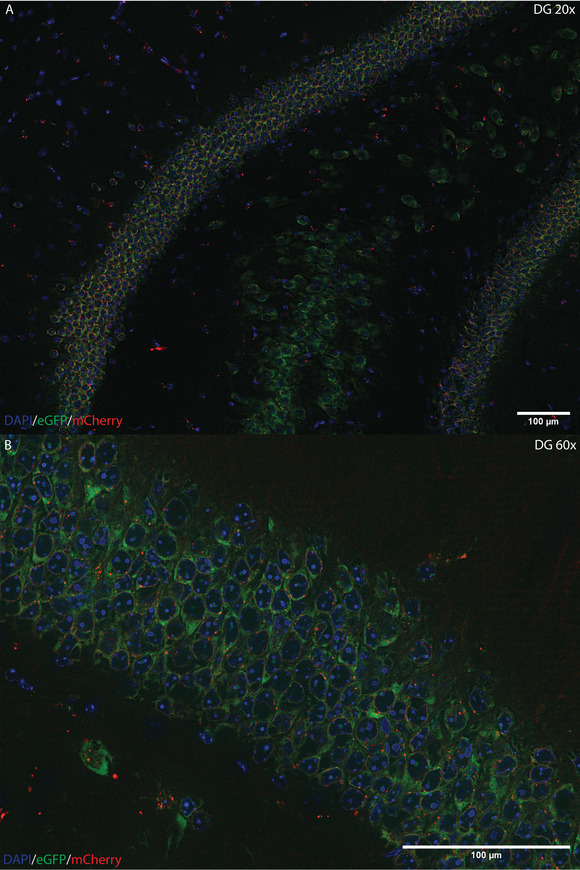
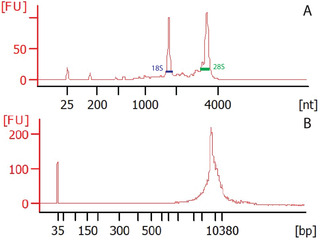
Epigenetic regulation of transcription is gaining increasing importance in the study of neurobiology. The advent of sequencing technology has enabled the study of this regulation across the entire genome and transcriptome. However, modern methods that allow the correlation of transcriptomic data with epigenomic regulation have had several key limitations, including use of separate tissue sources and detection of low-expression genes. This article describes a method combining isolation of nuclei tagged in specific cell types (INTACT) with translating ribosome affinity purification (TRAP) in the same cell homogenate, referred to as Simultaneous INTACT and TRAP (SIT). We used this technical approach to directly couple transcriptomic sequencing with epigenomic data in neurons derived from the mouse hippocampus. We demonstrate this method with an Emx1-NuTRAP transgenic mouse model. Here, we present protocols for SIT and for the generation and validation of the Emx1-NuTRAP mouse model that we used to demonstrate SIT. These methods enable cell type-specific comparison of translating mRNA and chromatin data from the same set of cells. Using SIT and the Emx1-NuTRAP transgenic mouse model, researchers can compare epigenomic data to transcriptomic data in the same set of hippocampal excitatory neurons. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC. Basic Protocol 1: Emx1-NuTRAP transgenic mouse line for labeling excitatory neurons in the hippocampus Basic Protocol 2: SIT: Simultaneous Isolation of nuclei tagged in specific cell types (INTACT) and Translating ribosome affinity purification (TRAP).
“SIT”与Emx1-NuTRAP小鼠:同时完整和陷阱配对转录组和表观遗传测序。
转录的表观遗传调控在神经生物学研究中越来越重要。测序技术的出现使得对整个基因组和转录组的这种调控的研究成为可能。然而,允许转录组数据与表观基因组调控相关联的现代方法有几个关键的局限性,包括使用单独的组织来源和检测低表达基因。本文介绍了一种在同一细胞匀浆中分离特定细胞类型标记的细胞核(完好无损)和翻译核糖体亲和纯化(TRAP)相结合的方法,称为同时完好无损和TRAP (SIT)。我们使用这种技术方法直接将转录组测序与小鼠海马神经元的表观基因组数据结合起来。我们用Emx1-NuTRAP转基因小鼠模型验证了这种方法。在这里,我们提出了SIT的协议,以及我们用来演示SIT的Emx1-NuTRAP小鼠模型的生成和验证。这些方法能够对来自同一组细胞的翻译mRNA和染色质数据进行细胞类型特异性比较。利用SIT和Emx1-NuTRAP转基因小鼠模型,研究人员可以比较同一组海马兴奋性神经元的表观基因组数据和转录组数据。©2022作者。当前协议由Wiley期刊有限责任公司发表。基本协议1:Emx1-NuTRAP转基因小鼠系标记海马兴奋性神经元基本协议2:SIT:同时分离特定细胞类型标记的细胞核(完好无损)和翻译核糖体亲和纯化(TRAP)。
本文章由计算机程序翻译,如有差异,请以英文原文为准。




 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


