RNA Methylome Reveals the m6A-mediated Regulation of Flavor Metabolites in Tea Leaves under Solar-withering
IF 7.9
2区 生物学
Q1 GENETICS & HEREDITY
引用次数: 0
Abstract
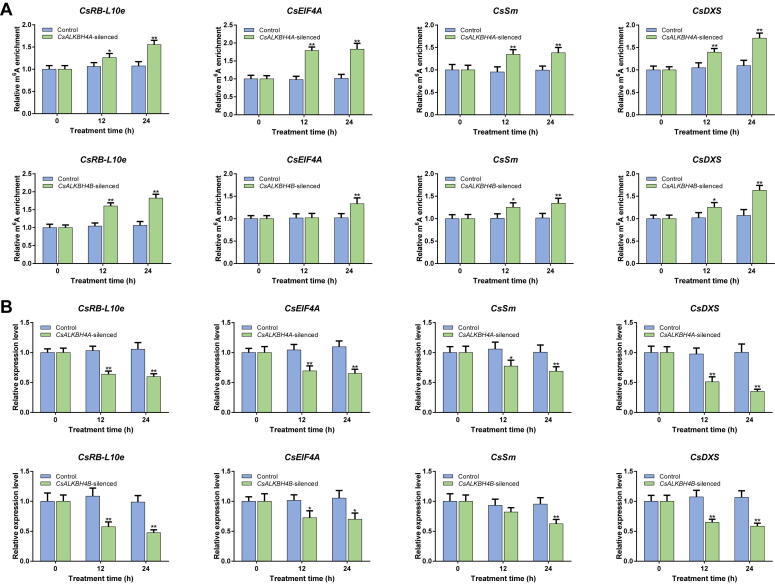
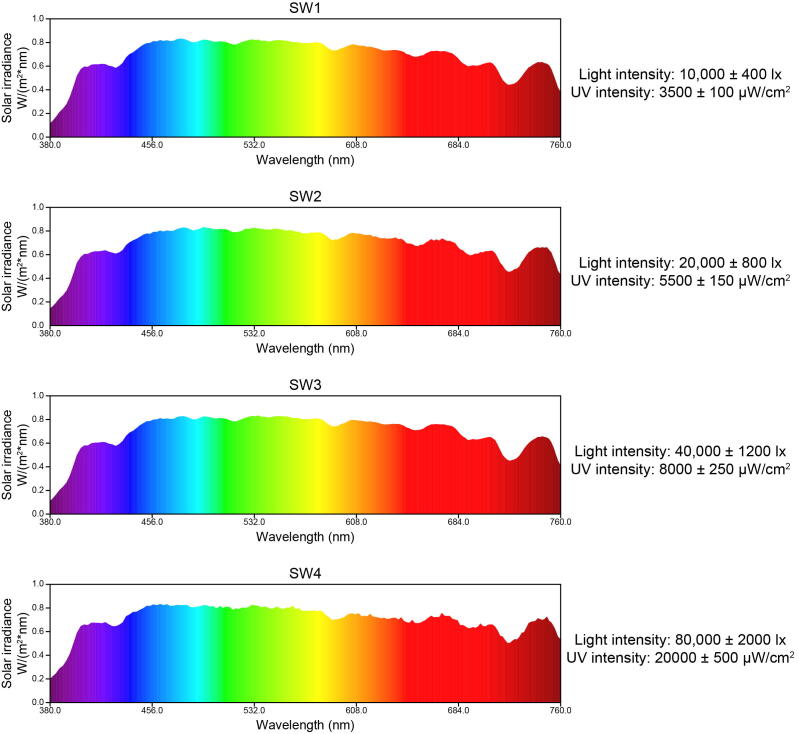
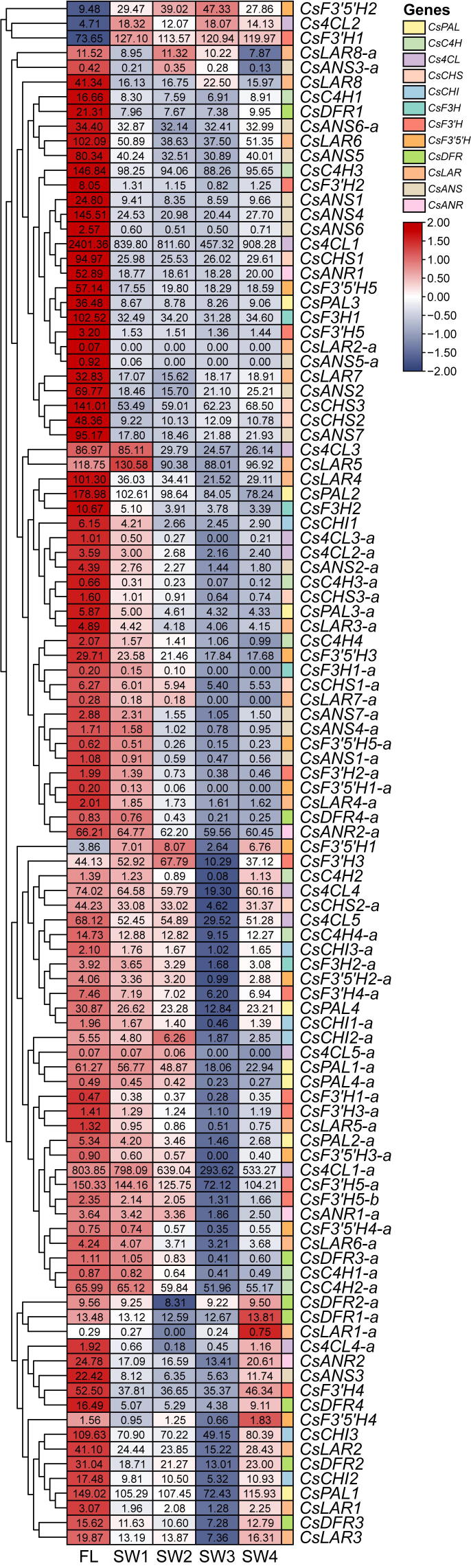
The epitranscriptomic mark N6-methyladenosine (m6A), which is the predominant internal modification in RNA, is important for plant responses to diverse stresses. Multiple environmental stresses caused by the tea-withering process can greatly influence the accumulation of specialized metabolites and the formation of tea flavor. However, the effects of the m6A-mediated regulatory mechanism on flavor-related metabolic pathways in tea leaves remain relatively uncharacterized. We performed an integrated RNA methylome and transcriptome analysis to explore the m6A-mediated regulatory mechanism and its effects on flavonoid and terpenoid metabolism in tea (Camellia sinensis) leaves under solar-withering conditions. Dynamic changes in global m6A level in tea leaves were mainly controlled by two m6A erasers (CsALKBH4A and CsALKBH4B) during solar-withering treatments. Differentially methylated peak-associated genes following solar-withering treatments with different shading rates were assigned to terpenoid biosynthesis and spliceosome pathways. Further analyses indicated that CsALKBH4-driven RNA demethylation can directly affect the accumulation of volatile terpenoids by mediating the stability and abundance of terpenoid biosynthesis-related transcripts and also indirectly influence the flavonoid, catechin, and theaflavin contents by triggering alternative splicing-mediated regulation. Our findings revealed a novel layer of epitranscriptomic gene regulation in tea flavor-related metabolic pathways and established a link between the m6A-mediated regulatory mechanism and the formation of tea flavor under solar-withering conditions.
RNA甲基组揭示m6a介导的日光萎蔫下茶叶风味代谢物调控
外转录组标记n6 -甲基腺苷(n6 - methylladenosine, m6A)是RNA中主要的内部修饰,在植物对各种逆境的响应中起着重要作用。茶叶萎凋过程中产生的多种环境胁迫对茶叶特殊代谢物的积累和茶叶风味的形成有很大的影响。然而,m6a介导的调节机制对茶叶风味相关代谢途径的影响尚不明确。我们通过RNA甲基组和转录组分析,探讨了m6a介导的调控机制及其对太阳萎蔫条件下茶(Camellia sinensis)叶片黄酮和萜类代谢的影响。日光枯萎处理期间,全球茶叶中m6A水平的动态变化主要受CsALKBH4A和CsALKBH4B两种m6A擦除剂控制。在不同遮光率的日光枯萎处理下,不同甲基化的峰相关基因与萜类生物合成和剪接体途径有关。进一步分析表明,csalkbh4驱动的RNA去甲基化可以通过介导萜类生物合成相关转录物的稳定性和丰度直接影响挥发性萜类的积累,也可以通过触发选择性剪接介导的调节间接影响类黄酮、儿茶素和茶黄素的含量。我们的研究结果揭示了一层新的表观转录组基因调控茶叶风味相关代谢途径,并建立了m6a介导的调控机制与日光萎凋条件下茶叶风味形成之间的联系。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Genomics, Proteomics & Bioinformatics
Biochemistry, Genetics and Molecular Biology-Biochemistry
CiteScore
14.30
自引率
4.20%
发文量
844
审稿时长
61 days
期刊介绍:
Genomics, Proteomics and Bioinformatics (GPB) is the official journal of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China. It aims to disseminate new developments in the field of omics and bioinformatics, publish high-quality discoveries quickly, and promote open access and online publication. GPB welcomes submissions in all areas of life science, biology, and biomedicine, with a focus on large data acquisition, analysis, and curation. Manuscripts covering omics and related bioinformatics topics are particularly encouraged. GPB is indexed/abstracted by PubMed/MEDLINE, PubMed Central, Scopus, BIOSIS Previews, Chemical Abstracts, CSCD, among others.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


