Frequency of change determines effectiveness of microbial response strategies
IF 10.8
1区 环境科学与生态学
Q1 ECOLOGY
引用次数: 0
Abstract
Nature challenges microbes with change at different frequencies and demands an effective response for survival. Here, we used controlled laboratory experiments to investigate the effectiveness of different response strategies, such as post-translational modification, transcriptional regulation, and specialized versus adaptable metabolisms. For this, we inoculated replicated chemostats with an enrichment culture obtained from sulfidic stream microbiomes 16 weeks prior. The chemostats were submitted to alternatingly oxic and anoxic conditions at three frequencies, with periods of 1, 4 and 16 days. The microbial response was recorded with 16S rRNA gene amplicon sequencing, shotgun metagenomics, transcriptomics and proteomics. Metagenomics resolved provisional genomes of all abundant bacterial populations, mainly affiliated with Proteobacteria and Bacteroidetes. Almost all these populations maintained a steady growth rate under both redox conditions at all three frequencies of change. Our results supported three conclusions: (1) Oscillating oxic/anoxic conditions selected for generalistic species, rather than species specializing in only a single condition. (2) A high frequency of change selected for strong codon usage bias. (3) Alignment of transcriptomes and proteomes required multiple generations and was dependent on a low frequency of change.
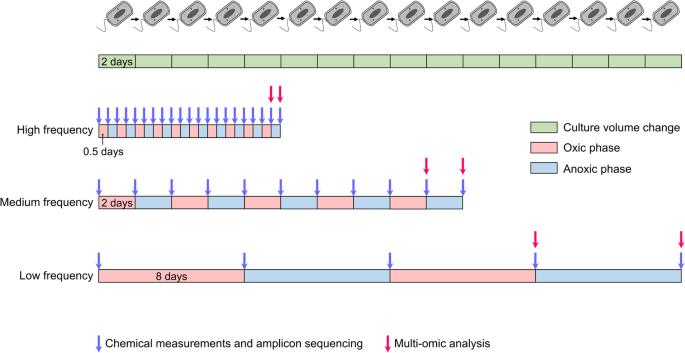
变化的频率决定了微生物反应策略的有效性。
大自然以不同频率的变化挑战微生物,并需要有效的生存反应。在这里,我们使用对照实验室实验来研究不同反应策略的有效性,如翻译后修饰、转录调控以及专门与适应性代谢。为此,我们用16周前从硫化物流微生物群中获得的富集培养物接种了复制的化学抑制剂。化学抑制剂在三个频率下交替置于有氧和缺氧条件下,周期分别为1、4和16天。用16S rRNA基因扩增子测序、鸟枪宏基因组学、转录组学和蛋白质组学记录微生物反应。宏基因组学解析了所有丰富细菌种群的临时基因组,主要隶属于变形菌门和拟杆菌门。几乎所有这些种群在两种氧化还原条件下,在所有三种变化频率下都保持稳定的增长率。我们的研究结果支持了三个结论:(1)为一般物种选择的振荡好氧/缺氧条件,而不是仅针对单一条件的物种。(2) 为强烈的密码子使用偏好而选择的高频率变化。(3) 转录组和蛋白质组的比对需要多代,并且依赖于低频率的变化。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

ISME Journal
环境科学-生态学
CiteScore
22.10
自引率
2.70%
发文量
171
审稿时长
2.6 months
期刊介绍:
The ISME Journal covers the diverse and integrated areas of microbial ecology. We encourage contributions that represent major advances for the study of microbial ecosystems, communities, and interactions of microorganisms in the environment. Articles in The ISME Journal describe pioneering discoveries of wide appeal that enhance our understanding of functional and mechanistic relationships among microorganisms, their communities, and their habitats.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


