Translocation of linearized full-length proteins through an engineered nanopore under opposing electrophoretic force
IF 33.1
1区 生物学
Q1 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
Nanopores have recently been used to identify and fingerprint proteins. However, because proteins, unlike DNA, do not have a uniform charge, the electrophoretic force cannot in general be used to translocate or linearize them. Here we show that the introduction of sets of charges in the lumen of the CytK nanopore spaced by ~1 nm creates an electroosmotic flow that induces the unidirectional transport of unstructured natural polypeptides against a strong electrophoretic force. Molecular dynamics simulations indicate that this electroosmotic-dominated force has a strength of ~20 pN at −100 mV, which is similar to the electric force on single-stranded DNA. Unfolded polypeptides produce current signatures as they traverse the nanopore, which may be used to identify proteins. This approach can be used to translocate and stretch proteins for enzymatic and non-enzymatic protein identification and sequencing. An engineered nanopore translocates untagged full-length proteins by electroosmotic force.
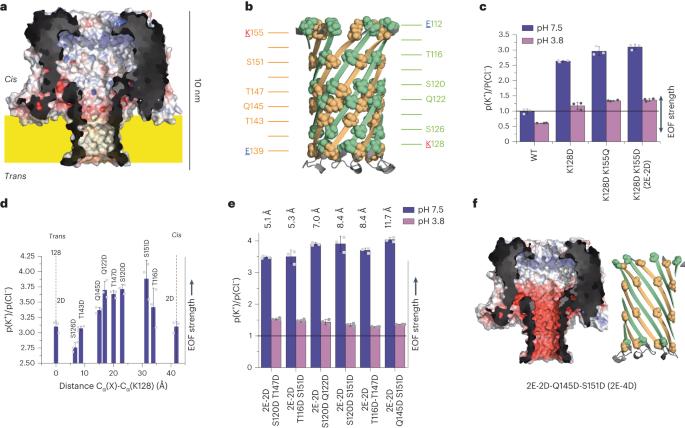
线性化的全长蛋白质在相反的电泳力下通过工程纳米孔的易位。
纳米孔最近被用于鉴定和指纹蛋白质。然而,由于蛋白质与DNA不同,没有均匀的电荷,电泳力通常不能用于易位或线性化。在这里,我们展示了在CytK纳米孔的内腔中引入的电荷集,间隔为~1 nm产生电渗流,该电渗流诱导非结构化天然多肽抵抗强电泳力的单向转运。分子动力学模拟表明,这种电渗主导的力的强度约为20 pN在-100 mV,这类似于单链DNA上的电力。未折叠的多肽在穿过纳米孔时产生电流特征,可用于识别蛋白质。该方法可用于易位和拉伸蛋白质,用于酶促和非酶促蛋白质鉴定和测序。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊
Nature biotechnology
工程技术-生物工程与应用微生物
CiteScore
63.00
自引率
1.70%
发文量
382
审稿时长
3 months
期刊介绍:
Nature Biotechnology is a monthly journal that focuses on the science and business of biotechnology. It covers a wide range of topics including technology/methodology advancements in the biological, biomedical, agricultural, and environmental sciences. The journal also explores the commercial, political, ethical, legal, and societal aspects of this research.
The journal serves researchers by providing peer-reviewed research papers in the field of biotechnology. It also serves the business community by delivering news about research developments. This approach ensures that both the scientific and business communities are well-informed and able to stay up-to-date on the latest advancements and opportunities in the field.
Some key areas of interest in which the journal actively seeks research papers include molecular engineering of nucleic acids and proteins, molecular therapy, large-scale biology, computational biology, regenerative medicine, imaging technology, analytical biotechnology, applied immunology, food and agricultural biotechnology, and environmental biotechnology.
In summary, Nature Biotechnology is a comprehensive journal that covers both the scientific and business aspects of biotechnology. It strives to provide researchers with valuable research papers and news while also delivering important scientific advancements to the business community.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


