DCAlign v1.0: aligning biological sequences using co-evolution models and informed priors.
IF 5.4
3区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
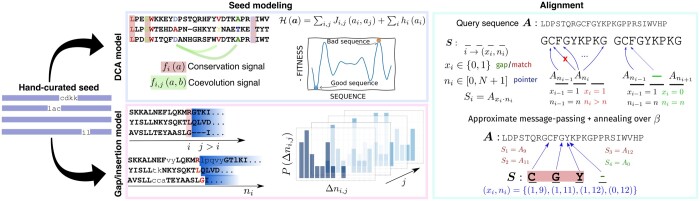
Summary: DCAlign is a new alignment method able to cope with the conservation and the co-evolution signals that characterize the columns of multiple sequence alignments of homologous sequences. However, the pre-processing steps required to align a candidate sequence are computationally demanding. We show in v1.0 how to dramatically reduce the overall computing time by including an empirical prior over an informative set of variables mirroring the presence of insertions and deletions.
Availability and implementation: DCAlign v1.0 is implemented in Julia and it is fully available at https://github.com/infernet-h2020/DCAlign.
DCAlign v1.0:使用协同进化模型和知情先验对生物序列进行对齐。
摘要:DCAlign是一种新的比对方法,能够处理同源序列多序列比对列的保守性和协同进化信号。然而,对齐候选序列所需的预处理步骤在计算上要求很高。我们在v1.0中展示了如何通过在反映插入和删除存在的一组信息变量上包含经验先验来显著减少总体计算时间。可用性和实现:DCAlign v1.0是在Julia中实现的,可以在https://github.com/infernet-h2020/DCAlign上完全获得。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Bioinformatics
生物-生化研究方法
CiteScore
11.20
自引率
5.20%
发文量
753
审稿时长
2.1 months
期刊介绍:
The leading journal in its field, Bioinformatics publishes the highest quality scientific papers and review articles of interest to academic and industrial researchers. Its main focus is on new developments in genome bioinformatics and computational biology. Two distinct sections within the journal - Discovery Notes and Application Notes- focus on shorter papers; the former reporting biologically interesting discoveries using computational methods, the latter exploring the applications used for experiments.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


