Non-steroidal anti-inflammatory drug target gene associations with major depressive disorders: a Mendelian randomisation study integrating GWAS, eQTL and mQTL Data
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 0
Abstract
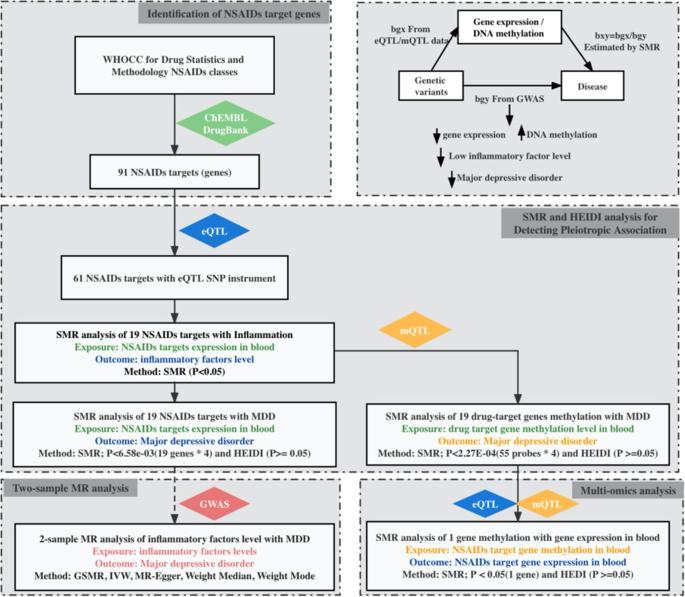
Previous observational studies reported associations between non-steroidal anti-inflammatory drugs (NSAIDs) and major depressive disorder (MDD), however, these associations are often inconsistent and underlying biological mechanisms are still poorly understood. We conducted a two-sample Mendelian randomisation (MR) study to examine relationships between genetic variants and NSAID target gene expression or DNA methylation (DNAm) using publicly available expression, methylation quantitative trait loci (eQTL or mQTL) data and genetic variant-disease associations from genome-wide association studies (GWAS of MDD). We also assessed drug exposure using gene expression and DNAm levels of NSAID targets as proxies. Genetic variants were robustly adjusted for multiple comparisons related to gene expression, DNAm was used as MR instrumental variables and GWAS statistics of MDD as the outcome. A 1-standard deviation (SD) lower expression of NEU1 in blood was related to lower C-reactive protein (CRP) levels of −0.215 mg/L (95% confidence interval (CI): 0.128–0.426) and a decreased risk of MDD (odds ratio [OR] = 0.806; 95% CI: 0.735–0.885; p = 5.36 × 10−6). A concordant direction of association was also observed for NEU1 DNAm levels in blood and a risk of MDD (OR = 0.886; 95% CI: 0.836–0.939; p = 4.71 × 10−5). Further, the genetic variants associated with MDD were mediated by NEU1 expression via DNAm (β = −0.519; 95% CI: −0.717 to −0.320256; p = 3.16 × 10−7). We did not observe causal relationships between inflammatory genetic marker estimations and MDD risk. Yet, we identified a concordant association of NEU1 messenger RNA and an adverse direction of association of higher NEU1 DNAm with MDD risk. These results warrant increased pharmacovigilance and further in vivo or in vitro studies to investigate NEU1 inhibitors or supplements for MDD.
非甾体抗炎药靶基因与重度抑郁障碍的关系:一项整合了 GWAS、eQTL 和 mQTL 数据的孟德尔随机化研究
以往的观察性研究报告了非甾体抗炎药(NSAIDs)与重度抑郁症(MDD)之间的关联,然而,这些关联往往并不一致,而且人们对其背后的生物学机制仍然知之甚少。我们进行了一项双样本孟德尔随机化(MR)研究,利用可公开获得的表达、甲基化定量性状位点(eQTL 或 mQTL)数据以及全基因组关联研究(GWAS of MDD)中的遗传变异-疾病关联,研究遗传变异与非甾体抗炎药靶基因表达或 DNA 甲基化(DNAm)之间的关系。我们还使用非甾体抗炎药靶点的基因表达和DNAm水平作为替代物评估了药物暴露。基因变异经稳健调整后用于与基因表达相关的多重比较,DNAm被用作MR工具变量,MDD的GWAS统计数据被用作结果。血液中 NEU1 表达量减少 1 个标准差 (SD) 与 C 反应蛋白 (CRP) 水平降低 -0.215 mg/L(95% 置信区间 (CI):0.128-0.426)和 MDD 风险降低有关(比值比 [OR] = 0.806;95% CI:0.735-0.885;P = 5.36 × 10-6)。血液中 NEU1 DNAm 水平与 MDD 风险的关联方向也一致(OR = 0.886;95% CI:0.836-0.939;p = 4.71 × 10-5)。此外,与 MDD 相关的遗传变异是通过 DNAm 的 NEU1 表达介导的(β = -0.519;95% CI:-0.717 至 -0.320256;p = 3.16 × 10-7)。我们没有观察到炎症遗传标记估计值与 MDD 风险之间的因果关系。然而,我们发现 NEU1 信使 RNA 与 MDD 风险之间存在一致的关联,而较高的 NEU1 DNAm 与 MDD 风险之间存在不利的关联方向。这些结果表明,有必要加强药物警戒,并进一步开展体内或体外研究,以调查治疗 MDD 的 NEU1 抑制剂或补充剂。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


