Prediction of mechanistic subtypes of Parkinson’s using patient-derived stem cell models
IF 18.8
1区 计算机科学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
Abstract
Parkinson’s disease is a common, incurable neurodegenerative disorder that is clinically heterogeneous: it is likely that different cellular mechanisms drive the pathology in different individuals. So far it has not been possible to define the cellular mechanism underlying the neurodegenerative disease in life. We generated a machine learning-based model that can simultaneously predict the presence of disease and its primary mechanistic subtype in human neurons. We used stem cell technology to derive control or patient-derived neurons, and generated different disease subtypes through chemical induction or the presence of mutation. Multidimensional fluorescent labelling of organelles was performed in healthy control neurons and in four different disease subtypes, and both the quantitative single-cell fluorescence features and the images were used to independently train a series of classifiers to build deep neural networks. Quantitative cellular profile-based classifiers achieve an accuracy of 82%, whereas image-based deep neural networks predict control and four distinct disease subtypes with an accuracy of 95%. The machine learning-trained classifiers achieve their accuracy across all subtypes, using the organellar features of the mitochondria with the additional contribution of the lysosomes, confirming the biological importance of these pathways in Parkinson’s. Altogether, we show that machine learning approaches applied to patient-derived cells are highly accurate at predicting disease subtypes, providing proof of concept that this approach may enable mechanistic stratification and precision medicine approaches in the future. Deep learning applied to live-cell images of patient-derived neurons aids predicting underlying mechanisms and gains insights into neurodegenerative diseases, facilitating the understanding of mechanistic heterogeneity. D’Sa and colleagues use patient-derived stem cell models, high-throughput imaging and machine learning algorithms to investigate Parkinson’s disease subtyping.
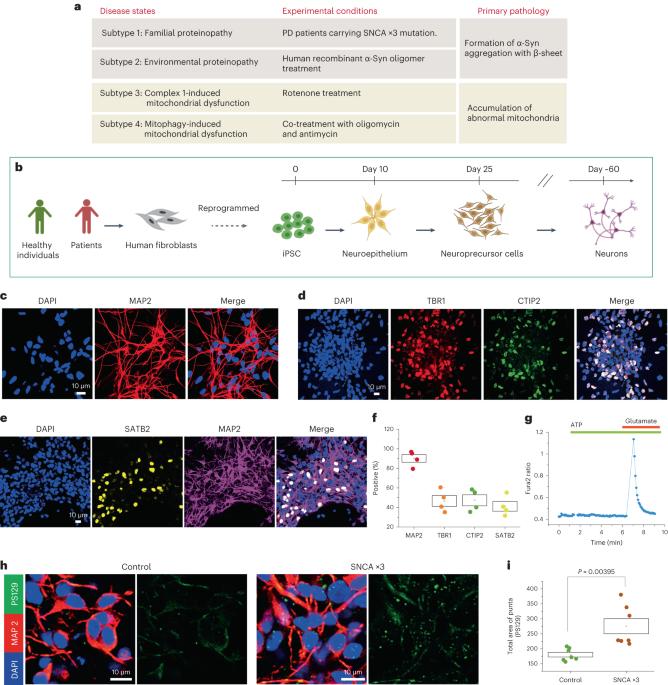
使用患者来源的干细胞模型预测帕金森氏症的机制亚型。
帕金森病是一种常见的、无法治愈的神经退行性疾病,具有临床异质性:可能是不同的细胞机制驱动了不同个体的病理。到目前为止,还无法确定生活中神经退行性疾病的细胞机制。我们生成了一个基于机器学习的模型,可以同时预测人类神经元中疾病及其主要机制亚型的存在。我们使用干细胞技术衍生出对照或患者衍生的神经元,并通过化学诱导或突变产生不同的疾病亚型。在健康对照神经元和四种不同的疾病亚型中进行细胞器的多维荧光标记,并使用定量单细胞荧光特征和图像独立训练一系列分类器以构建深度神经网络。基于定量细胞图谱的分类器实现了82%的准确率,而基于图像的深度神经网络预测控制和四种不同的疾病亚型的准确率为95%。机器学习训练的分类器利用线粒体的器质特征和溶酶体的额外贡献,在所有亚型中实现了准确性,证实了这些途径在帕金森氏症中的生物学重要性。总之,我们表明,应用于患者衍生细胞的机器学习方法在预测疾病亚型方面非常准确,这为这种方法在未来实现机制分层和精确医学方法提供了概念证明。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Machine Intelligence
Multiple-
CiteScore
36.90
自引率
2.10%
发文量
127
期刊介绍:
Nature Machine Intelligence is a distinguished publication that presents original research and reviews on various topics in machine learning, robotics, and AI. Our focus extends beyond these fields, exploring their profound impact on other scientific disciplines, as well as societal and industrial aspects. We recognize limitless possibilities wherein machine intelligence can augment human capabilities and knowledge in domains like scientific exploration, healthcare, medical diagnostics, and the creation of safe and sustainable cities, transportation, and agriculture. Simultaneously, we acknowledge the emergence of ethical, social, and legal concerns due to the rapid pace of advancements.
To foster interdisciplinary discussions on these far-reaching implications, Nature Machine Intelligence serves as a platform for dialogue facilitated through Comments, News Features, News & Views articles, and Correspondence. Our goal is to encourage a comprehensive examination of these subjects.
Similar to all Nature-branded journals, Nature Machine Intelligence operates under the guidance of a team of skilled editors. We adhere to a fair and rigorous peer-review process, ensuring high standards of copy-editing and production, swift publication, and editorial independence.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


