XGDAG: explainable gene-disease associations via graph neural networks.
IF 5.4
3区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
Abstract
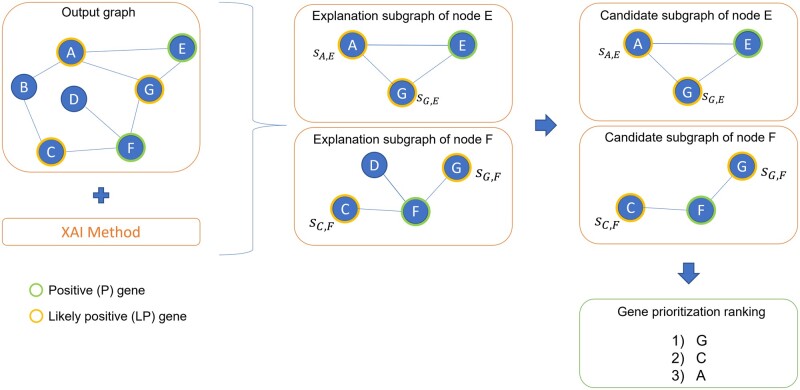
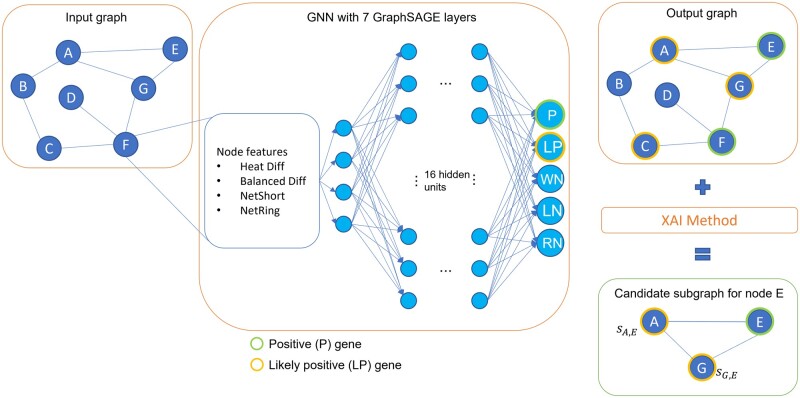
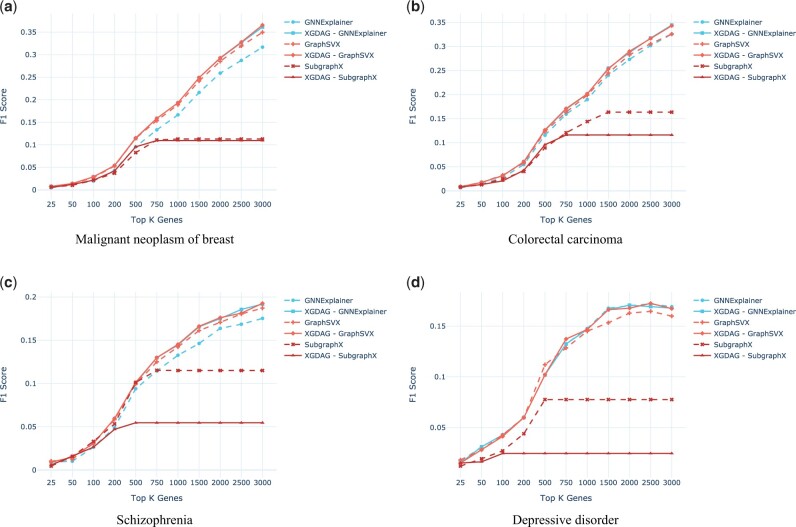
Abstract Motivation Disease gene prioritization consists in identifying genes that are likely to be involved in the mechanisms of a given disease, providing a ranking of such genes. Recently, the research community has used computational methods to uncover unknown gene–disease associations; these methods range from combinatorial to machine learning-based approaches. In particular, during the last years, approaches based on deep learning have provided superior results compared to more traditional ones. Yet, the problem with these is their inherent black-box structure, which prevents interpretability. Results We propose a new methodology for disease gene discovery, which leverages graph-structured data using graph neural networks (GNNs) along with an explainability phase for determining the ranking of candidate genes and understanding the model’s output. Our approach is based on a positive–unlabeled learning strategy, which outperforms existing gene discovery methods by exploiting GNNs in a non-black-box fashion. Our methodology is effective even in scenarios where a large number of associated genes need to be retrieved, in which gene prioritization methods often tend to lose their reliability. Availability and implementation The source code of XGDAG is available on GitHub at: https://github.com/GiDeCarlo/XGDAG. The data underlying this article are available at: https://www.disgenet.org/, https://thebiogrid.org/, https://doi.org/10.1371/journal.pcbi.1004120.s003, and https://doi.org/10.1371/journal.pcbi.1004120.s004.
XGDAG:通过图神经网络解释基因与疾病的关联。
动机:疾病基因优先排序包括识别可能参与特定疾病机制的基因,并对这些基因进行排序。最近,研究界使用计算方法来揭示未知的基因与疾病的关联;这些方法包括从组合到基于机器学习的方法。特别是,在过去的几年里,基于深度学习的方法比传统的方法提供了更好的结果。然而,它们的问题在于它们固有的黑箱结构,这阻碍了可解释性。结果:我们提出了一种疾病基因发现的新方法,该方法利用图神经网络(gnn)的图结构数据以及可解释性阶段来确定候选基因的排名并理解模型的输出。我们的方法基于一种积极的无标签学习策略,该策略通过以非黑箱方式利用gnn,优于现有的基因发现方法。即使在需要检索大量相关基因的情况下,我们的方法也是有效的,在这种情况下,基因优先排序方法往往会失去其可靠性。可用性和实现:XGDAG的源代码可在GitHub上获得:https://github.com/GiDeCarlo/XGDAG。本文的基础数据可在以下位置获得:https://www.disgenet.org/、https://thebiogrid.org/、https://doi.org/10.1371/journal.pcbi.1004120.s003和https://doi.org/10.1371/journal.pcbi.1004120.s004。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Bioinformatics
生物-生化研究方法
CiteScore
11.20
自引率
5.20%
发文量
753
审稿时长
2.1 months
期刊介绍:
The leading journal in its field, Bioinformatics publishes the highest quality scientific papers and review articles of interest to academic and industrial researchers. Its main focus is on new developments in genome bioinformatics and computational biology. Two distinct sections within the journal - Discovery Notes and Application Notes- focus on shorter papers; the former reporting biologically interesting discoveries using computational methods, the latter exploring the applications used for experiments.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


