CYP2C19 代谢状态对 SSRI 反应的影响:对 9500 名澳大利亚抑郁症遗传学研究参与者的回顾性研究
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 13
摘要
CYP2C19 基因的变异与选择性血清素再摄取抑制剂(SSRIs)的不同代谢有关。基于 CYP2C19 变异影响的药物遗传学建议已经问世,并被越来越多的临床医师采用。然而,将不同代谢与疗效或不良副作用联系起来的基本假设仍未得到充分研究。在此,我们以 9531 名服用过 SSRIs 的澳大利亚成年人为样本,研究他们的 CYP2C19 多态性和推断代谢以及患者报告的抗抑郁药反应,旨在填补这一空白。根据 CYP2C19 等位基因推断参与者的代谢状态。主要分析包括评估正常代谢者(参照)与超快速、快速、中等和不良代谢者之间在疗效和耐受性方面的差异。在所有药物中,代谢差者的疗效较高,而代谢快者的耐受性较高。如果按药物进行分层,代谢状态与疗效之间的联系经不起多重测试校正。中度代谢者报告舍曲林有任何副作用的几率更高,报告的各种药物和舍曲林副作用的次数也更多。代谢状态与疗效、耐受性和副作用之间的影响与预期方向一致。我们的功率分析表明,我们可以检测到中等至较大的效应,至少是名义上的效应。我们没有测量抗抑郁药剂量或同时服用药物的异质性,这也可能是导致功率下降的原因。即使不调整临床滴定,我们也发现代谢较慢的患者出现副作用的风险较高,而且名义上的显著关联与预期的代谢效应一致,这为CYP2C19代谢与SSRI反应之间的联系提供了新的证据。尽管如此,要确定CYP2C19代谢对SSRI疗效或不良反应的影响,还需要纵向和干预性设计,如按代谢状态分层的随机临床试验。本文章由计算机程序翻译,如有差异,请以英文原文为准。
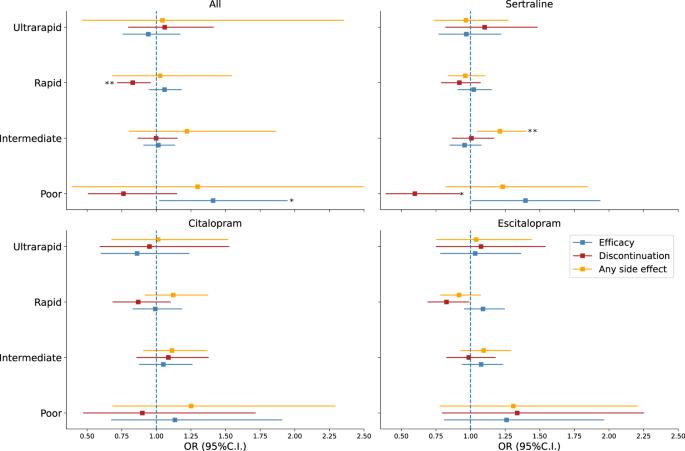
Impact of CYP2C19 metaboliser status on SSRI response: a retrospective study of 9500 participants of the Australian Genetics of Depression Study
Variation within the CYP2C19 gene has been linked to differential metabolism of selective serotonin reuptake inhibitors (SSRIs). Pharmacogenetic recommendations based on the effect of CYP2C19 variants have been made available and are used increasingly by clinical practitioners. Nonetheless, the underlying assumption linking differential metabolism to efficacy or adverse side effects remains understudied. Here, we aim to fill this gap by studying CYP2C19 polymorphisms and inferred metabolism and patient-reported antidepressant response in a sample of 9531 Australian adults who have taken SSRIs. Metaboliser status was inferred for participants based on CYP2C19 alleles. Primary analysis consisted of assessing differences in treatment efficacy and tolerability between normal (reference) and: ultrarapid, rapid, intermediate and poor metabolisers. Across medications, poor metabolisers reported a higher efficacy, whereas rapid metabolisers reported higher tolerability. When stratified by drug, associations between metaboliser status and efficacy did not survive multiple testing correction. Intermediate metabolisers were at greater odds of reporting any side effect for sertraline and higher number of side effects across medications and for sertraline. The effects between metaboliser status and treatment efficacy, tolerability and side effects were in the expected direction. Our power analysis suggests we would detect moderate to large effects, at least nominally. Reduced power may also be explained by heterogeneity in antidepressant dosages or concomitant medications, which we did not measure. The fact that we identify slower metabolisers to be at higher risk of side effects even without adjusting for clinical titration, and the nominally significant associations consistent with the expected metabolic effects provide new evidence for the link between CYP2C19 metabolism and SSRI response. Nonetheless, longitudinal and interventional designs such as randomized clinical trials that stratify by metaboliser status are necessary to establish the effects of CYP2C19 metabolism on SSRI treatment efficacy or adverse effects.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


