Tom van den Ende, Ymke van der Pol, Aafke Creemers, Norbert Moldovan, Dries Boers, Mark I van Berge Henegouwen, Maarten CCM Hulshof, Saskia AGM Cillessen, Nicole CT van Grieken, D Michiel Pegtel, Sarah Derks, Maarten F Bijlsma, Florent Mouliere, Hanneke WM van Laarhoven
下载PDF
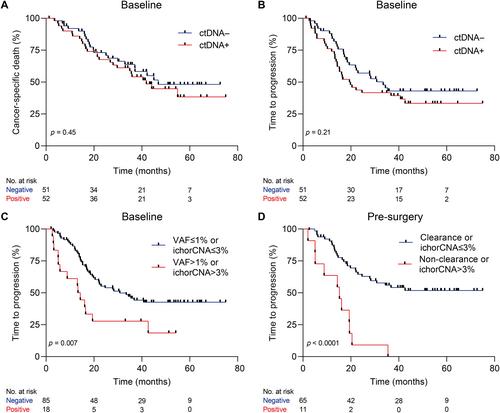
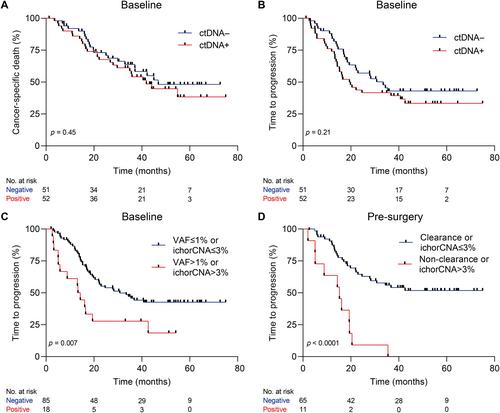
{"title":"可切除食管腺癌患者的全基因组和基于面板的无细胞DNA特征","authors":"Tom van den Ende, Ymke van der Pol, Aafke Creemers, Norbert Moldovan, Dries Boers, Mark I van Berge Henegouwen, Maarten CCM Hulshof, Saskia AGM Cillessen, Nicole CT van Grieken, D Michiel Pegtel, Sarah Derks, Maarten F Bijlsma, Florent Mouliere, Hanneke WM van Laarhoven","doi":"10.1002/path.6175","DOIUrl":null,"url":null,"abstract":"<p>Circulating tumor DNA (ctDNA) holds promise in resectable esophageal adenocarcinoma (EAC) to predict patient outcome but is not yet sensitive enough to be clinically applicable. Our aim was to combine ctDNA mutation data with shallow whole-genome sequencing (sWGS)-derived copy number tumor fraction estimates (ichorCNA) to improve pathological response and survival prediction in EAC. In total, 111 stage II/III EAC patients with baseline (<i>n</i> = 111), post-neoadjuvant chemoradiotherapy (nCRT) (<i>n</i> = 68), and pre-surgery (<i>n</i> = 92) plasma samples were used for ctDNA characterization. sWGS (<5× coverage) was performed on all time-point samples, and copy number aberrations were estimated using ichorCNA. Baseline and pre-surgery samples were sequenced using a custom amplicon panel for mutation detection. Detection of baseline ctDNA was successful in 44.3% of patients by amplicon sequencing and 10.5% by ichorCNA. Combining both, ctDNA could be detected in 50.5% of patients. Baseline ctDNA positivity was related to higher T stage (cT3, 4) (<i>p</i> = 0.017). There was no relationship between pathological response and baseline ctDNA positivity. However, baseline ctDNA metrics (variant allele frequency > 1% or ichorCNA > 3%) were associated with a high risk of disease progression [HR = 2.23 (95% CI 1.22–4.07), <i>p</i> = 0.007]. The non-clearance of a baseline variant or ichorCNA > 3% in pre-surgery samples was related to early progression [HR = 4.58 (95% CI 2.22–9.46), <i>p</i> < 0.001]. Multi-signal analysis improves detection of ctDNA and can be used for prognostication of resectable EAC patients. Future studies should explore the potential of multi-modality sequencing for risk stratification and treatment adaptation based on ctDNA results. © 2023 The Authors. <i>The Journal of Pathology</i> published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.</p>","PeriodicalId":232,"journal":{"name":"The Journal of Pathology","volume":"261 3","pages":"286-297"},"PeriodicalIF":5.6000,"publicationDate":"2023-08-24","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://onlinelibrary.wiley.com/doi/epdf/10.1002/path.6175","citationCount":"0","resultStr":"{\"title\":\"Genome-wide and panel-based cell-free DNA characterization of patients with resectable esophageal adenocarcinoma\",\"authors\":\"Tom van den Ende, Ymke van der Pol, Aafke Creemers, Norbert Moldovan, Dries Boers, Mark I van Berge Henegouwen, Maarten CCM Hulshof, Saskia AGM Cillessen, Nicole CT van Grieken, D Michiel Pegtel, Sarah Derks, Maarten F Bijlsma, Florent Mouliere, Hanneke WM van Laarhoven\",\"doi\":\"10.1002/path.6175\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>Circulating tumor DNA (ctDNA) holds promise in resectable esophageal adenocarcinoma (EAC) to predict patient outcome but is not yet sensitive enough to be clinically applicable. Our aim was to combine ctDNA mutation data with shallow whole-genome sequencing (sWGS)-derived copy number tumor fraction estimates (ichorCNA) to improve pathological response and survival prediction in EAC. In total, 111 stage II/III EAC patients with baseline (<i>n</i> = 111), post-neoadjuvant chemoradiotherapy (nCRT) (<i>n</i> = 68), and pre-surgery (<i>n</i> = 92) plasma samples were used for ctDNA characterization. sWGS (<5× coverage) was performed on all time-point samples, and copy number aberrations were estimated using ichorCNA. Baseline and pre-surgery samples were sequenced using a custom amplicon panel for mutation detection. Detection of baseline ctDNA was successful in 44.3% of patients by amplicon sequencing and 10.5% by ichorCNA. Combining both, ctDNA could be detected in 50.5% of patients. Baseline ctDNA positivity was related to higher T stage (cT3, 4) (<i>p</i> = 0.017). There was no relationship between pathological response and baseline ctDNA positivity. However, baseline ctDNA metrics (variant allele frequency > 1% or ichorCNA > 3%) were associated with a high risk of disease progression [HR = 2.23 (95% CI 1.22–4.07), <i>p</i> = 0.007]. The non-clearance of a baseline variant or ichorCNA > 3% in pre-surgery samples was related to early progression [HR = 4.58 (95% CI 2.22–9.46), <i>p</i> < 0.001]. Multi-signal analysis improves detection of ctDNA and can be used for prognostication of resectable EAC patients. Future studies should explore the potential of multi-modality sequencing for risk stratification and treatment adaptation based on ctDNA results. © 2023 The Authors. <i>The Journal of Pathology</i> published by John Wiley & Sons Ltd on behalf of The Pathological Society of Great Britain and Ireland.</p>\",\"PeriodicalId\":232,\"journal\":{\"name\":\"The Journal of Pathology\",\"volume\":\"261 3\",\"pages\":\"286-297\"},\"PeriodicalIF\":5.6000,\"publicationDate\":\"2023-08-24\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://onlinelibrary.wiley.com/doi/epdf/10.1002/path.6175\",\"citationCount\":\"0\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"The Journal of Pathology\",\"FirstCategoryId\":\"3\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/path.6175\",\"RegionNum\":2,\"RegionCategory\":\"医学\",\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"Q1\",\"JCRName\":\"ONCOLOGY\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"The Journal of Pathology","FirstCategoryId":"3","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/path.6175","RegionNum":2,"RegionCategory":"医学","ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"ONCOLOGY","Score":null,"Total":0}
引用次数: 0
引用
批量引用

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


