Gene expression signatures of human cell and tissue longevity
IF 4.1
Q2 GERIATRICS & GERONTOLOGY
引用次数: 42
Abstract
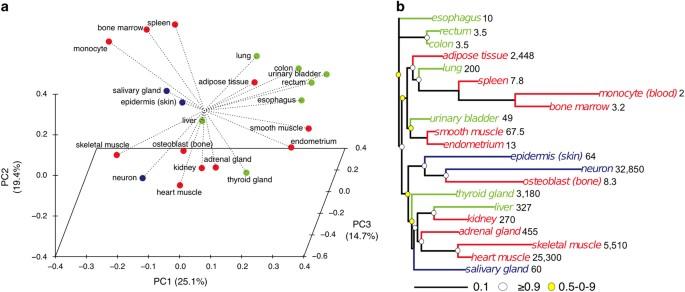
Different cell types within the body exhibit substantial variation in the average time they live, ranging from days to the lifetime of the organism. The underlying mechanisms governing the diverse lifespan of different cell types are not well understood. To examine gene expression strategies that support the lifespan of different cell types within the human body, we obtained publicly available RNA-seq data sets and interrogated transcriptomes of 21 somatic cell types and tissues with reported cellular turnover, a bona fide estimate of lifespan, ranging from 2 days (monocytes) to a lifetime (neurons). Exceptionally long-lived neurons presented a gene expression profile of reduced protein metabolism, consistent with neuronal survival and similar to expression patterns induced by longevity interventions such as dietary restriction. Across different cell lineages, we identified a gene expression signature of human cell and tissue turnover. In particular, turnover showed a negative correlation with the energetically costly cell cycle and factors supporting genome stability, concomitant risk factors for aging-associated pathologies. In addition, the expression of p53 was negatively correlated with cellular turnover, suggesting that low p53 activity supports the longevity of post-mitotic cells with inherently low risk of developing cancer. Our results demonstrate the utility of comparative approaches in unveiling gene expression differences among cell lineages with diverse cell turnover within the same organism, providing insights into mechanisms that could regulate cell longevity. Human tissue and cell types exhibit different gene signatures based on their cellular lifespans. Vadim Gladyshev and colleagues from Brigham and Women’s Hospital, Harvard Medical School, analyzed the gene expression patterns of 21 different cell types with cellular turnover times ranging from 2 days (white blood cells) to a lifetime (neurons). This turnover–defined as the balance between cell proliferation and death–has been shown to be a good estimate of cellular lifespan. The authors found that long-lived cell lineages, including those of the muscle and the brain, showed lower expression of genes involved in promoting cell division and maintaining genomic fidelity, consistent with a molecular path toward longevity. The finding that cells use lineage-specific strategies to alter their lifespans lays the groundwork for future therapies that promote human longevity by modifying gene expression profiles.
人类细胞和组织长寿的基因表达特征
人体内不同类型细胞的平均寿命有很大差异,从几天到整个生物体的一生。目前还不太清楚支配不同细胞类型不同寿命的内在机制。为了研究支持人体内不同细胞类型寿命的基因表达策略,我们获得了可公开获得的 RNA-seq 数据集,并对 21 种体细胞类型和组织的转录组进行了研究,这些细胞类型和组织的细胞周转率(一种对寿命的真实估计)从 2 天(单核细胞)到终生(神经元)不等。异常长寿的神经元呈现出蛋白质代谢减少的基因表达谱,这与神经元的存活一致,也与饮食限制等长寿干预措施诱导的表达模式相似。在不同的细胞系中,我们发现了人类细胞和组织更替的基因表达特征。特别是,新陈代谢与能量消耗高的细胞周期和支持基因组稳定性的因素呈负相关,而这些因素都是衰老相关病症的风险因素。此外,p53 的表达与细胞更替呈负相关,这表明低 p53 活性支持有丝分裂后细胞的长寿,其罹患癌症的风险本身就很低。我们的研究结果证明了比较方法在揭示同一生物体内不同细胞更替的细胞系之间基因表达差异方面的实用性,为了解细胞长寿的调控机制提供了启示。人类组织和细胞类型根据其细胞寿命表现出不同的基因特征。哈佛大学医学院布里格姆妇女医院的瓦迪姆-格拉迪舍夫及其同事分析了21种不同细胞类型的基因表达模式,这些细胞类型的细胞更替时间从2天(白细胞)到一生(神经元)不等。这种周转被定义为细胞增殖和死亡之间的平衡,已被证明是对细胞寿命的良好估计。作者发现,包括肌肉和大脑在内的长寿细胞系中,参与促进细胞分裂和维持基因组保真度的基因表达较低,这与长寿的分子途径一致。这一发现为未来通过改变基因表达谱来促进人类长寿的疗法奠定了基础。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


