LETTER TO THE EDITOR Performance of the ViroSeq® HIV-1 Genotyping System v2.0 in Central Africa.
The Open AIDS Journal
Pub Date : 2015-02-27
eCollection Date: 2015-01-01
DOI:10.2174/1874613601509010009
引用次数: 7
Abstract
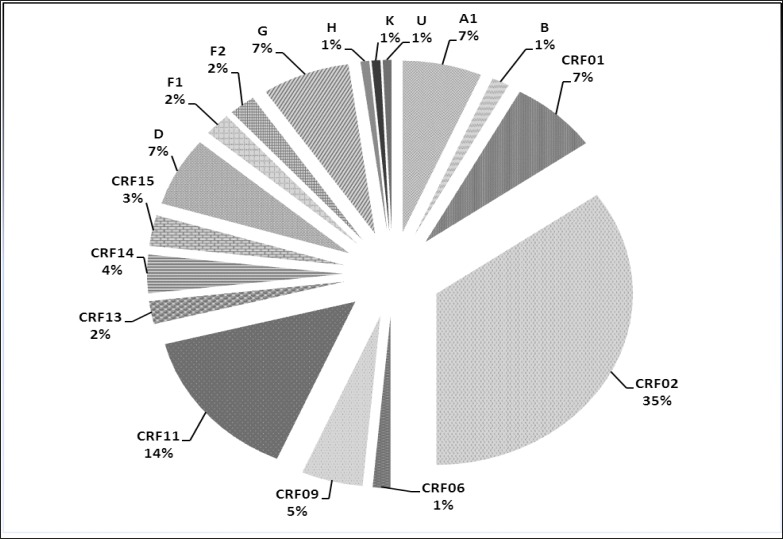
Resistance genotypes in pol gene of HIV-1 were obtained by the ViroSeq(®) HIV-1 Genotyping System v2.0 (Celera Diagnostics, Alameda, CA, USA) in 138 of 145 (95%) antiretroviral treatment-experienced adults in virological failure living in Central Africa (Cameroon, Central African Republic, Chad, Gabon). HIV-1 group M exhibited broad genetic diversity. Performance of the 7 ViroSeq(®) sequencing primers showed high failure rate, from 3% to 76% (D: 76%; F: 17%; A and H: 15%; G and B: 4%; C: 3%). These findings emphasize the need of updating the ViroSeq(®) HIV-1 genotyping system for non-B subtypes HIV-1.
致编辑:ViroSeq®HIV-1基因分型系统v2.0在中非的表现
利用ViroSeq(®)HIV-1基因分型系统v2.0 (Celera Diagnostics, Alameda, CA, USA)对生活在中非(喀麦隆、中非共和国、乍得、加蓬)145名(95%)接受过抗逆转录病毒治疗且病毒学失败的成人中的138人进行了HIV-1 pol基因耐药基因型分析。HIV-1 M组表现出广泛的遗传多样性。7种ViroSeq(®)测序引物的性能显示出高失败率,从3%到76% (D: 76%;F: 17%;A和H: 15%;G和B: 4%;C: 3%)。这些发现强调了更新针对非b亚型HIV-1的ViroSeq(®)HIV-1基因分型系统的必要性。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


