A multi-modal single-cell and spatial expression map of metastatic breast cancer biopsies across clinicopathological features
IF 58.7
1区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
Abstract
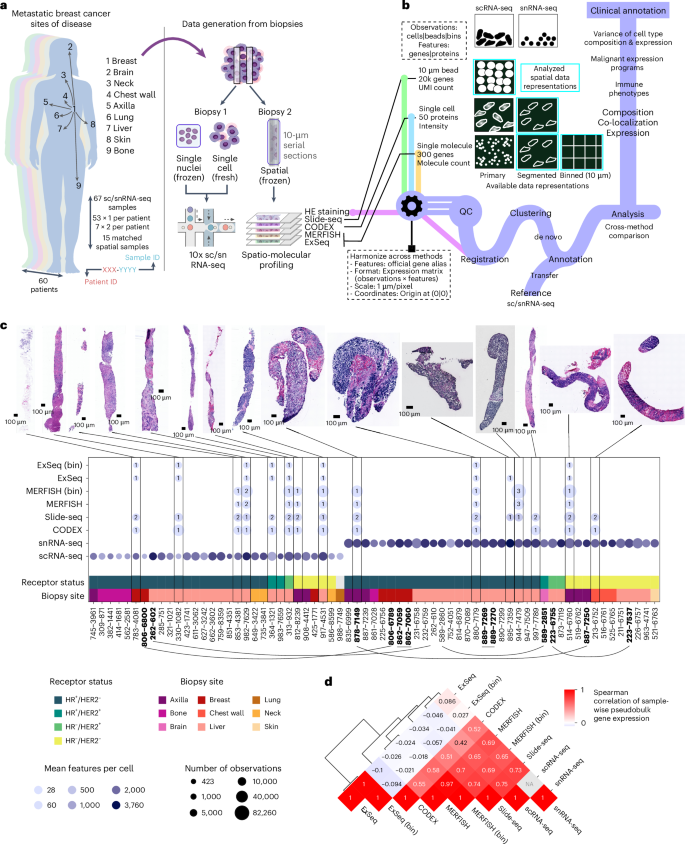
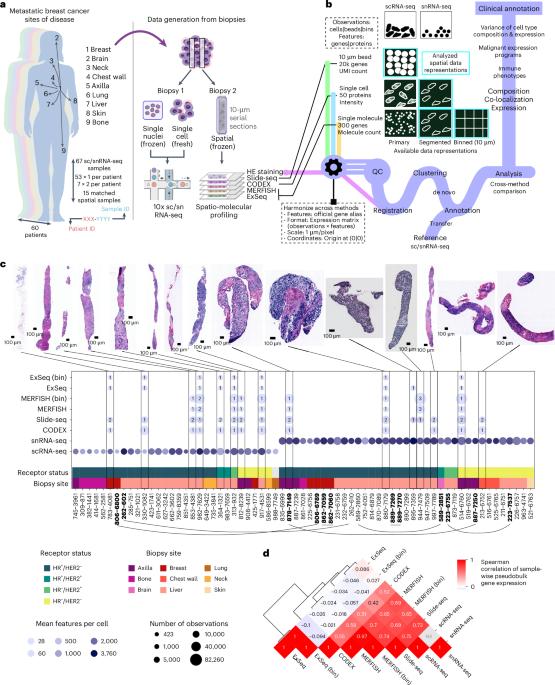
Although metastatic disease is the leading cause of cancer-related deaths, its tumor microenvironment remains poorly characterized due to technical and biospecimen limitations. In this study, we assembled a multi-modal spatial and cellular map of 67 tumor biopsies from 60 patients with metastatic breast cancer across diverse clinicopathological features and nine anatomic sites with detailed clinical annotations. We combined single-cell or single-nucleus RNA sequencing for all biopsies with a panel of four spatial expression assays (Slide-seq, MERFISH, ExSeq and CODEX) and H&E staining of consecutive serial sections from up to 15 of these biopsies. We leveraged the coupled measurements to provide reference points for the utility and integration of different experimental techniques and used them to assess variability in cell type composition and expression as well as emerging spatial expression characteristics across clinicopathological and methodological diversity. Finally, we assessed spatial expression and co-localization features of macrophage populations, characterized three distinct spatial phenotypes of epithelial-to-mesenchymal transition and identified expression programs associated with local T cell infiltration versus exclusion, showcasing the potential of clinically relevant discovery in such maps. Single-nucleus and single-cell RNA sequencing plus spatial profiling with four methods of core biopsies from 60 patients with metastatic breast cancer reveal patient-specific gene expression programs of breast cancer metastases that are maintained across time, site of metastasis and spatial profiling method, with spatial phenotypes correlating with microenvironmental features.
跨临床病理特征的转移性乳腺癌活组织多模式单细胞和空间表达图谱
尽管转移性疾病是癌症相关死亡的主要原因,但由于技术和生物样本的限制,其肿瘤微环境的特征仍然不甚明了。在这项研究中,我们对来自 60 名转移性乳腺癌患者的 67 份肿瘤活检样本进行了多模态空间和细胞图谱分析,这些活检样本具有不同的临床病理特征和九个解剖部位,并附有详细的临床注释。我们将所有活检样本的单细胞或单核 RNA 测序与四种空间表达检测方法(Slide-seq、MERFISH、ExSeq 和 CODEX)组合在一起,并对其中多达 15 个活检样本的连续序列切片进行 H&E 染色。我们利用耦合测量为不同实验技术的实用性和整合提供参考点,并利用它们评估细胞类型组成和表达的变异性,以及临床病理学和方法学多样性中新出现的空间表达特征。最后,我们评估了巨噬细胞群的空间表达和共定位特征,描述了上皮向间质转化的三种不同空间表型,并确定了与局部 T 细胞浸润和排斥相关的表达程序,展示了在此类图谱中发现临床相关信息的潜力。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Nature Medicine
医学-生化与分子生物学
CiteScore
100.90
自引率
0.70%
发文量
525
审稿时长
1 months
期刊介绍:
Nature Medicine is a monthly journal publishing original peer-reviewed research in all areas of medicine. The publication focuses on originality, timeliness, interdisciplinary interest, and the impact on improving human health. In addition to research articles, Nature Medicine also publishes commissioned content such as News, Reviews, and Perspectives. This content aims to provide context for the latest advances in translational and clinical research, reaching a wide audience of M.D. and Ph.D. readers. All editorial decisions for the journal are made by a team of full-time professional editors.
Nature Medicine consider all types of clinical research, including:
-Case-reports and small case series
-Clinical trials, whether phase 1, 2, 3 or 4
-Observational studies
-Meta-analyses
-Biomarker studies
-Public and global health studies
Nature Medicine is also committed to facilitating communication between translational and clinical researchers. As such, we consider “hybrid” studies with preclinical and translational findings reported alongside data from clinical studies.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


