Investigation of plasmid-mediated quinolone resistance among extended-spectrum β-lactamase isolates of E. coli and K. pneumoniae
IF 2.1
4区 医学
Q3 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
Abstract
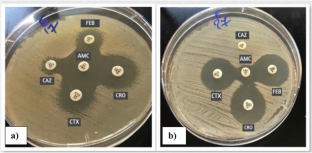
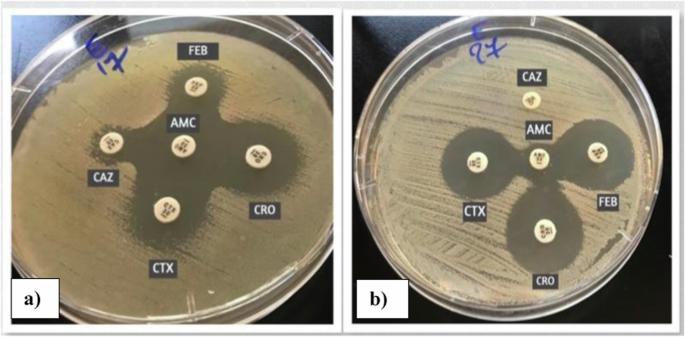
Escherichia coli and Klebsiella pneumoniae are important members of the Enterobacteriaceae family, involved in many infections. The increased resistance rate towards β-lactams and fluoroquinolones -which are the main therapeutic options- limits their treatment options. This study aimed to assess the local resistance patterns against different antimicrobials and to determine the extended-spectrum β-lactamase (ESBLs) producers. The study revealed that 36% of clinical isolates were ESBL producers, showing high resistance rates towards β-lactams and non-β-lactams, especially sulphamethoxazole-trimethoprim and fluoroquinolones. However, they were susceptible to chloramphenicol and doxycycline (33% and 20%; respectively). Also, the investigation aimed to screen the plasmid profile of quinolone-resistant ESBLs-producers and to detect the plasmid-mediated quinolone resistance genes including qnrA, qnrS, qnrB, qnrC, qnrD, and qnrVC. Moreover, the conjugative plasmid among the quinolone-resistant isolates was elucidated. The results showed that extracted plasmids of sizes ranging from ≈0.9 to 21.23 Kb, divided into 7 plasmid patterns were detected. A plasmid of approximately 21.23 Kb was found in all isolates and the QnrS gene was the most predominant gene. Moreover, the frequency of transconjugation within the same genus was higher than that recorded between different genera; where 68% of E. coli isolates transferred the resistance genes compared to Klebsiella isolates (36.6%). Plasmid profiles of transconjugants demonstrated great similarity, where 21.23 Kb plasmid was detected in all transconjugants. Since these transconjugants were quinolone-resistant ESBL producers, it has been suggested that quinolone resistance determinants might be carried on that plasmid.
对大肠杆菌和肺炎双球菌的广谱β-内酰胺酶分离物中质粒介导的喹诺酮类药物耐药性的调查。
大肠埃希菌和肺炎克雷伯菌是肠杆菌科的重要成员,涉及多种感染。大肠埃希菌和肺炎克雷伯菌是肠杆菌科细菌中的重要成员,涉及多种感染,它们对β-内酰胺类药物和氟喹诺酮类药物(主要治疗选择)的耐药率增加,限制了它们的治疗选择。本研究旨在评估当地对不同抗菌素的耐药性模式,并确定广谱β-内酰胺酶(ESBLs)的产生者。研究发现,36% 的临床分离菌株是 ESBL 生产者,对 β-内酰胺类和非 β-内酰胺类药物,尤其是磺胺甲噁唑-三甲氧苄啶和氟喹诺酮类药物的耐药率很高。不过,它们对氯霉素和强力霉素敏感(分别为 33% 和 20%)。调查还旨在筛选耐喹诺酮 ESBLs 生产者的质粒特征,并检测质粒介导的喹诺酮耐药基因,包括 qnrA、qnrS、qnrB、qnrC、qnrD 和 qnrVC。此外,还阐明了耐喹诺酮分离物中的共轭质粒。结果表明,提取的质粒大小从≈0.9 Kb到21.23 Kb不等,分为7种质粒模式。在所有分离株中都发现了约 21.23 Kb 的质粒,QnrS 基因是最主要的基因。此外,同属内的转共轭频率高于不同属间的转共轭频率;68%的大肠杆菌分离物转移了抗性基因,而克雷伯氏菌分离物则为 36.6%。转共轭物的质粒图谱显示出很大的相似性,在所有转共轭物中都检测到了 21.23 Kb 的质粒。由于这些转座菌是耐奎诺酮的 ESBL 生产者,因此认为该质粒上可能携带有奎诺酮耐药基因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文
来源期刊

Journal of Antibiotics
医学-免疫学
CiteScore
6.60
自引率
3.00%
发文量
87
审稿时长
1 months
期刊介绍:
The Journal of Antibiotics seeks to promote research on antibiotics and related types of biologically active substances and publishes Articles, Review Articles, Brief Communication, Correspondence and other specially commissioned reports. The Journal of Antibiotics accepts papers on biochemical, chemical, microbiological and pharmacological studies. However, studies regarding human therapy do not fall under the journal’s scope. Contributions regarding recently discovered antibiotics and biologically active microbial products are particularly encouraged. Topics of particular interest within the journal''s scope include, but are not limited to, those listed below:
Discovery of new antibiotics and related types of biologically active substances
Production, isolation, characterization, structural elucidation, chemical synthesis and derivatization, biological activities, mechanisms of action, and structure-activity relationships of antibiotics and related types of biologically active substances
Biosynthesis, bioconversion, taxonomy and genetic studies on producing microorganisms, as well as improvement of production of antibiotics and related types of biologically active substances
Novel physical, chemical, biochemical, microbiological or pharmacological methods for detection, assay, determination, structural elucidation and evaluation of antibiotics and related types of biologically active substances
Newly found properties, mechanisms of action and resistance-development of antibiotics and related types of biologically active substances.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


