Zea mays genotype influences microbial and viral rhizobiome community structure
IF 5.1
Q1 ECOLOGY
引用次数: 0
Abstract
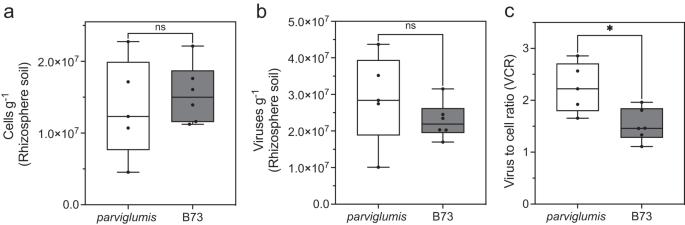
Plant genotype is recognized to contribute to variations in microbial community structure in the rhizosphere, soil adherent to roots. However, the extent to which the viral community varies has remained poorly understood and has the potential to contribute to variation in soil microbial communities. Here we cultivated replicates of two Zea mays genotypes, parviglumis and B73, in a greenhouse and harvested the rhizobiome (rhizoplane and rhizosphere) to identify the abundance of cells and viruses as well as rhizobiome microbial and viral community using 16S rRNA gene amplicon sequencing and genome resolved metagenomics. Our results demonstrated that viruses exceeded microbial abundance in the rhizobiome of parviglumis and B73 with a significant variation in both the microbial and viral community between the two genotypes. Of the viral contigs identified only 4.5% (n = 7) of total viral contigs were shared between the two genotypes, demonstrating that plants even at the level of genotype can significantly alter the surrounding soil viral community. An auxiliary metabolic gene associated with glycoside hydrolase (GH5) degradation was identified in one viral metagenome-assembled genome (vOTU) identified in the B73 rhizobiome infecting Propionibacteriaceae (Actinobacteriota) further demonstrating the viral contribution in metabolic potential for carbohydrate degradation and carbon cycling in the rhizosphere. This variation demonstrates the potential of plant genotype to contribute to microbial and viral heterogeneity in soil systems and harbors genes capable of contributing to carbon cycling in the rhizosphere.
玉米基因型可能影响微生物和病毒根瘤菌群落结构。
植物基因型被认为有助于根际微生物群落结构的变化,土壤对根的附着。然而,病毒群落的变化程度仍然知之甚少,并且有可能导致土壤微生物群落的变化。在此,我们在温室中培养了两个玉米基因型parviglumis和B73的重复,并收获了根瘤菌群(根际和根际),利用16S rRNA基因扩增子测序和基因组解析宏基因组学来鉴定细胞和病毒的丰度,以及根瘤菌群微生物和病毒群落。结果表明,病毒在细小菌和B73的根瘤菌群中丰度超过微生物,且两种基因型之间的微生物和病毒群落存在显著差异。在鉴定的病毒群中,只有4.5% (n = 7)的病毒群在两种基因型之间是共享的,这表明即使在基因型水平上,植物也可以显著改变周围土壤的病毒群落。在感染丙酸杆菌科(放线菌科)的B73根瘤菌群中发现的一个病毒宏基因组组装基因组(vOTU)中发现了一个与糖苷水解酶(GH5)降解相关的辅助代谢基因,进一步证明了病毒在根际碳水化合物降解和碳循环的代谢潜力中的贡献。这种变异表明,植物基因型可能导致土壤系统中微生物和病毒的异质性,并携带能够促进根际碳循环的基因。
本文章由计算机程序翻译,如有差异,请以英文原文为准。
求助全文
约1分钟内获得全文
求助全文

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


